Figure 3.
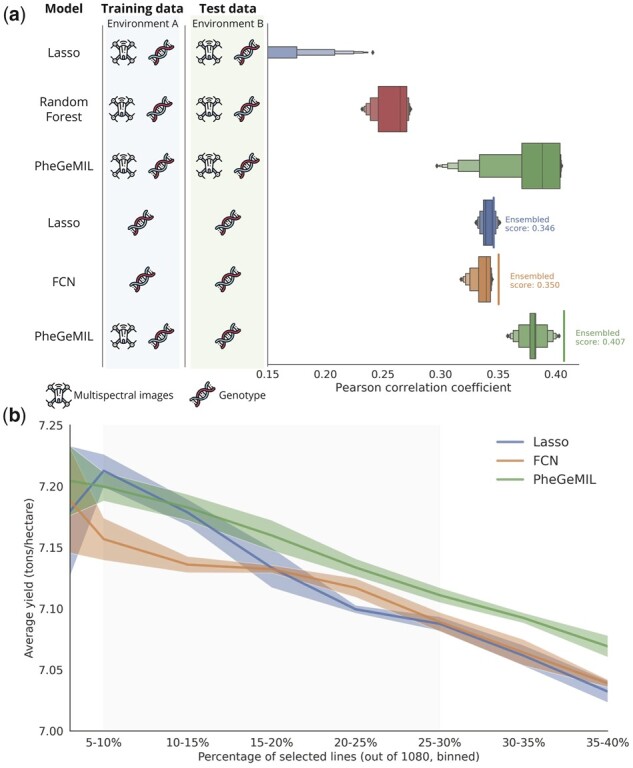
Generalization performance on a new environment and utilization for crop selection. (a) Comparison of yield prediction performance of a linear baseline (Lasso), two nonlinear baselines (Random forest and FCN) and our model (PheGeMIL) for prediction on a new, unseen environment using genotypic or phenotypic data. Multiple scenarios are evaluated. In all cases, training is done on data from environment A (2018 YT, see Table 1) and testing is done on data from environment B (2018 EYT). A set of experiments is conducted by training and evaluating on both multispectral images and genotypic data (first three rows). A second set of experiments is conducted by evaluating on genotypes alone (last three rows), to mimic prediction before sowing in breeding program scenarios. For baselines, training and testing must be done on the same data types and training can only be done on genotype alone. PheGeMIL, on the contrary, is trained with phenotypic data too, while still being evaluated on genotypes alone, thanks to the MIL framework. Distributions represent the performance in terms of Person correlation coefficient obtained on models trained on the 5 different splits of the training set. Ensembled performance for genotype-only predictions represents the prediction performance obtained when averaging the predicted values for a given sample across the 5 trained models. (b) Average yield obtained from a prediction-driven line selection of varying sizes (binned) using rankings derived from the values predicted by different ensembled methods. Lines are selected based on the predicted yield, their effective yield is then averaged across the set of selected lines and reported for an increasing selection size, ranging from 5% to 40% of the lines in the test set. MIL, multiple instance learning; FCN, fully connected network.
