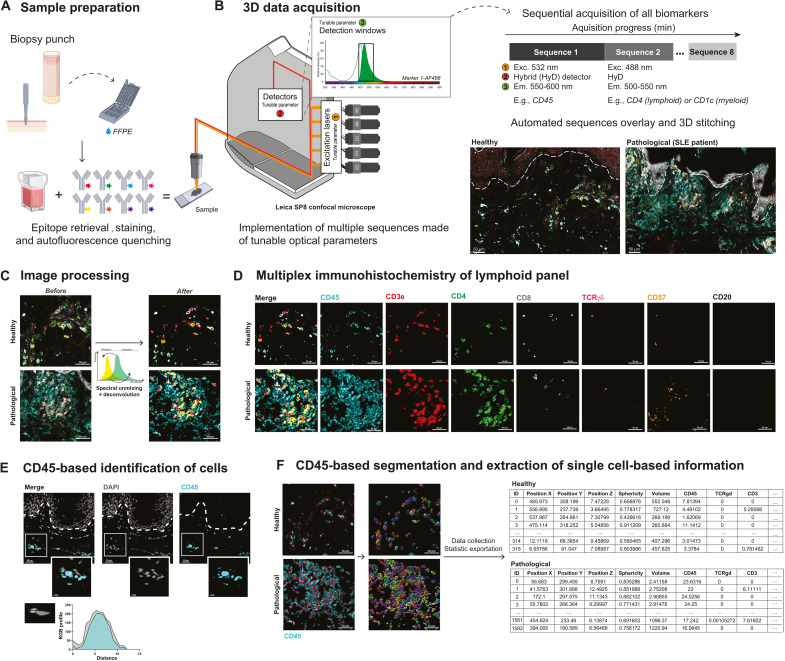
Fig. 1. Between-stack microscope configuration allows sequential acquisition of 7+ channels with classical image processing.
(A) Sample preparation. FFPE-skin sections were cut and stained for myeloid and lymphoid panels after appropriate epitope retrieval and autofluorescence quenching. Sample images were then acquired using an SP8 confocal microscope from Leica Microsystems as described in (B). (B) Microscope configuration and acquisition settings. Mosaic sequential images were acquired using the between-stack configuration with tunable detection windows. Sequences were overlaid and 3D-stitched. An example of data acquisition is given for healthy (left) and pathological [systemic lupus erythematosus (SLE)] (right) skin. (C) Deconvolution of regions of interest and spectral unmixing. Acquired 3D images were deconvoluted and compensated to correct optical aberrations and 3D fluorescent spectral spillovers. (D) Representative 3D multiplex image of healthy (top) and pathological SLE (bottom) skin sample for lymphoid panel, staining CD45, CD3, CD4, CD8, TCRγδ, CD57, and CD20. (E) Colocalization of DAPI (4′,6-diamidino-2-phenylindole) and CD45 staining and respective RGB profiles. (F) Segmentation and single-cell database creation. Cell segmentation using the CD45 fluorescence channel allowed efficient isolation of individual objects, i.e., immune cells. Individual object statistics (xyz coordinates, sphericity, volume, and MFI) were extracted for each sample. Scale bars, 30 μm.