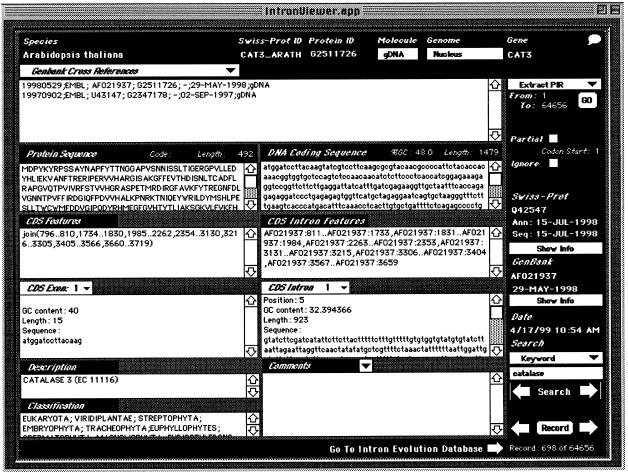
Figure 1.
Structure and Macintosh™ graphical user interface of the IDB. From left, top to bottom: fields are shown that identify the species, molecule, genome, and gene from which a particular nucleotide sequence is derived. SWISS-PROT Identification codes and the protein PID are also noted. Directly below the species and molecular identifiers is a list of all known database cross references followed by the coding region amino acid and DNA sequences. GC content and the length of particular molecules are noted where appropriate. The GenBank coding sequence join statement and derivative intron locations used to extract individual exon and intron sequences are available via pull-down menus. Position, phase, GC content, and sequence length are noted where appropriate. A description of the gene and the species classification taken from the GenBank Taxonomy database are also provided. The right panel contains additional information concerning the particular cross-references from SWISS-PROT and GenBank (including all accession numbers and database update dates) that were used to generate the database entry. The user may select from various navigational modes that allow examination of the database sequentially, by user-defined record, or by a search of genus, species, or protein description. User-defined ranges of records may be exported as text files for incorporation into other databases.