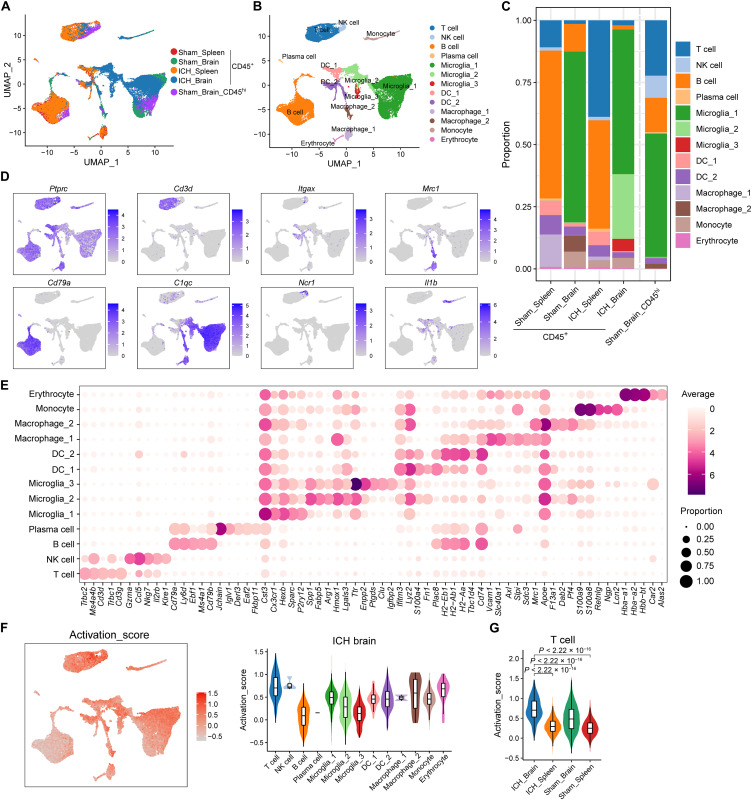
Fig. 3. Single-cell analysis of CD45+ cells isolated from the brain and spleen of ICH mice.
CD45+ cells were isolated from splenocytes or brain tissues of C57BL/6 mice 72 hours after autologous blood injection induced ICH using FACS. CD45+ cells splenic (Sham spleen) or brain (Sham brain_CD45+) cells from sham-operated mice were used as controls for unbiasedly single-cell RNA mapping. The optimized FACS sorting was performed on CD45hi cell population to obtain enough CD4+ T cells in sham brains for subclustering and data processing (Sham brain_ CD45hi). scRNA-seq was performed using the 10X Genomics Chromium platform. (A and B) Uniform manifold approximation and projection (UMAP) plot along components 1 and 2 for all high-quality single cells from sham brain_CD45+ (n = 10487 cells), sham brain_CD45hi (n = 5589 cells), sham spleen (n = 8747 cells), ICH brain (n = 10,487 cells), and ICH spleen (n = 9147 cells), colored by group and cell type. (C) Stacked bar chart showing the constitution of 13 cell clusters in sham spleen, sham brain_CD45+, sham brain_CD45hi, ICH spleen, and ICH brain. (D) UMAP plots show selected marker genes for determining cell identity. (E) Dot plots show average (dot color) and percentage (dot size) expression of top five marker genes for each cluster. (F) UMAP plot illustrating activation feature scores (Ms4a4b, Ms4a6b, Cxcl10, Itk, Prkcq, Ccnd1, Ccnd2, Ccnd3, Hmox1, Ccl5, Dusp1, Klf4, Jun, and Junb) calculated using AddModuleScore function in Seurat (left). Violin plot with box plot inside showing the comparison of activation scores between T cells and other CD3−CD45+ clusters in ICH Brain (right). (G) Violin plot with box plot inside illustrating the comparison of T cell activation scores between indicated groups. In (A) to (G), data were pooled from 15 mice in each group.