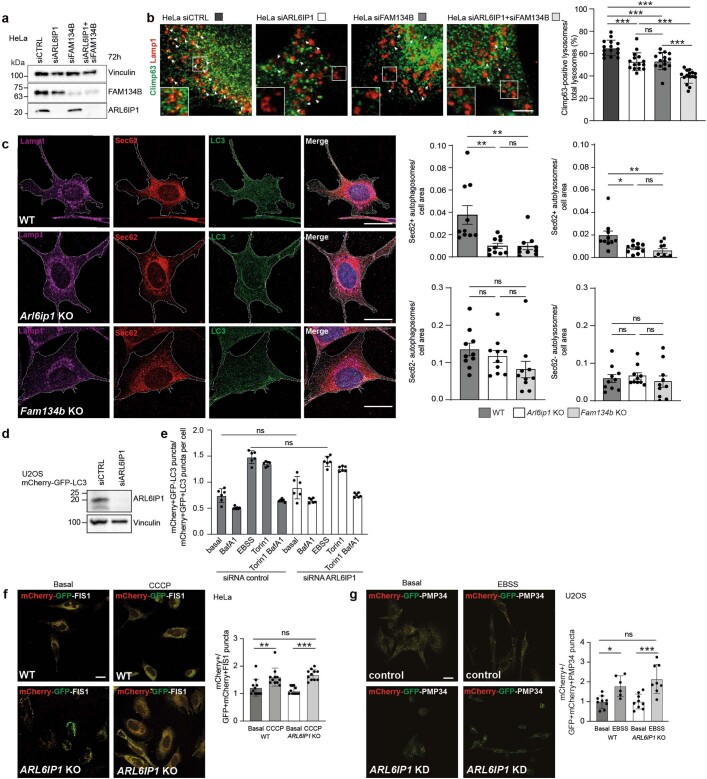
Extended Data Fig. 7. ER-phagy but not bulk autophagy, mitophagy or pexophagy is compromised upon disruption of ARL6IP1.
a) siRNA mediated knock-down of either ARL6IP1, FAM134B or both in HeLa cells (3 exp.). b) The ratio of Climp63-positive versus Climp63-negative lysosomes upon knock-down of either ARL6IP1 or FAM134B is consistent with a defect in ER-phagy. Cells were treated with Torin1 and Bafilomycin A1. The simultaneous knock-down of both further decreases this ratio (1 exp.; 15 cells per genotype, one-way-ANOVA, F-value = 32.59, p = 0.0001, Bonferroni post-hoc analysis: siCTRL versus siARL6IP1 p = 0.0001, siCTRL versus siFAM134B p = 0.0017, siRNA CTRL versus siARL6IP1+siFAM134B p = 0.0001; siARL6IP1 versus siARL6IP1+siFAM134B p = 0.0001, siFAM134B versus siARL6IP1+siFAM134B p = 0.0001). Scale bar: 5 µm. c) Arl6ip1 and Fam134b WT and KO MEFs were EBSS starved for 4 h, fixed and stained for Lamp1, LC3B, and the ER-protein Sec62. The quantification of autophagosomes (LC3B-positive and Lamp1-negative) loaded with ER (Sec62-positive) or devoid of ER (Sec62-negative) supports a defect in ER-phagy but not bulk autophagy (1 exp.; 10 cells per genotype were analysed; one-way-ANOVA with Bonferroni post-hoc analysis; autophagosomes: p = 0.001, F-value = 9.081, WT versus Arl6ip1 KO p = 0.003, WT versus Fam134b KO p = 0.003; autolysosomes: p = 0.004, F-value = 6.954, WT versus Arl6ip1 KO p = 0.024, WT versus Fam134b KO p = 0.005). Scale bar: 20 µm. d) Knock-down of ARL6IP1 in U2OS cells with inducible expression of the mCherry-GFP-LC3 reporter (1 exp.). e) Bulk autophagy upon siRNA mediated knock-down of ARL6IP1 is not compromised in U2OS cells expressing the mCherry-GFP-LC3 reporter (1 exp. with 3 replicates, one-way-ANOVA with Bonferroni post-hoc analysis). f) Mitophagy is not affected by KO of ARL6IP1. WT and ARL6IP1 KO HeLa cells were transfected with the mitophagy reporter mCherry-GFP-FIS1. Mitophagy flux was studied at steady state (basal) and after 4 h with 40 µM CCCP (1 exp; n = 11 cells each were analysed; one-way ANOVA with Bonferroni post-hoc analysis, F-value = 13.36, p = 0.0001: WT basal versus CCCP p = 0.0067, KO basal versus CCCP p = 0.0001). Scale bar: 25 µm. g) Pexophagy is not affected by ARL6IP1 knock-down. The mCherry-GFP-PMP34 reporter was induced in control and ARL6IP1 knock-down U2OS cells and pexophagy flux studied at steady state (basal) and 20 h of EBSS starvation (1 exp. with n = 9/6/9/8 cells analysed; one-way ANOVA with Bonferroni post-hoc analysis, F-value = 10.65, p = 0.0001: WT basal versus CCCP p = 0.0377, KO basal versus CCCP p = 0.0004). Scale bar: 25 µm. Quantitative data are shown as mean ± SEM.