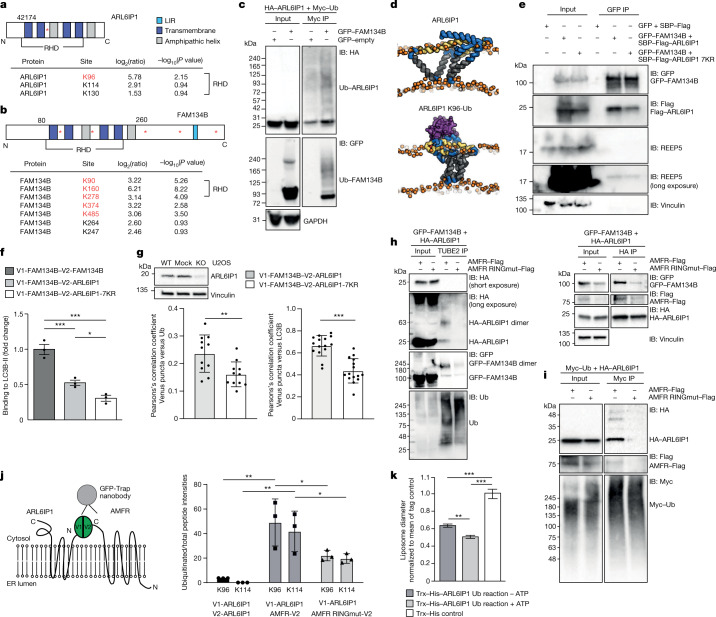
Fig. 4. LC3B binding to FAM134B–ARL6IP1 complexes depends on ARL6IP1 ubiquitination.
a,b, Position and log2(fold changes) of ubiquitinated lysine residues identified by LC–MS in ARL6IP1 (cells expressing V1-ARL6IP1–V2-ARL6IP1 or V1-FAM134B–V2-ARL6IP1) (a) and FAM134B (cells expressing V1-FAM134B–V2-FAM134B or V1-FAM134B–V2-ARL6IP1) (b). Significant sites are indicated in red (–log10(P value) > 1.3, one-sided unpaired Student’s t-test). c, Anti-Myc IP from HEK293T cells co-expressing HA–ARL6IP1, GFP–FAM134B and Myc–ubiquitin (Ub) confirms ARL6IP1 and FAM134B ubiquitination (one experiment). d, Snapshots from coarse-grained molecular dynamics simulations showing the most populated conformations of non-ubiquitinated and ubiquitinated ARL6IP1 (ubiquitin purple) in 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC 16:0/18:1) bilayers (orange). e, Interaction with FAM134B but not endogenous REEP5 is promoted by ARL6IP1 ubiquitination (two experiments). f, LC3B co-precipitation with V1-FAM134B–V2-ARL6IP1-7KR is reduced compared with V1-FAM134B–V2-ARL6IP1 (three experiments; one-way ANOVA, F = 46; Holm–Sidak’s post-test, FAM134B–FAM134B versus FAM134B–ARL6IP1, P = 0.0014; FAM134B–FAM134B versus FAM134B–ARL6IP1-7KR, P = 0.0002; FAM134B–ARL6IP1 versus FAM134B–ARL6IP1-7KR, P = 0.0242). g, Pearson’s correlation analysis for ubiquitin-positive and LC3B-positive Venus puncta for V1-FAM134B–V2-ARL6IP1 and V1-FAM134B–V2-ARL6IP1-7KR in ARL6IP1 KO U2OS cells (1 experiment with n = 11 (ubiquitin) and 15 (LC3B) cells; two-sided unpaired Student’s t-test, P = 0.006 (ubiquitin) and P = 0.0001 (LC3B)). h, Left, ubiquitinated ARL6IP1 is detected after TUBE2 pull-down after co-expression of GFP–FAM134B and HA–ARL6IP1 with AMFR–Flag but not AMFR-RINGmut–Flag. Right, ARL6IP1–FAM134B interaction is promoted by AMFR but not AMFR-RINGmut–Flag (n = 1). i, Myc pull-down after co-expression of Myc–ubiquitin–HA–ARL6IP1 with AMFR–Flag or AMFR-RINGmut–Flag confirms ARL6IP1 ubiquitination with active AMFR (n = 1). j, Schematic (left) and quantification (right) of LC–MS of GFP pull-downs of co-expressed V1-ARL6IP1 and AMFR-V2 or AMFR-RINGmut-V2: AMFR ubiquitinates ARL6IP1 at K96 and K114 (one-sided unpaired Student’s t-test: V1-ARL6IP1–V2-ARL6IP1 versus V1-ARL6IP1–AMFR-V2 K96, P = 0.0075; K114, P = 0.006; V1-ARL6IP1–AMFR-V2 versus V1-ARL6IP1–AMFR-RINGmut-V2 K96, P = 0.038; K114, P = 0.044). k, In vitro ubiquitination of Trx–His–ARL6IP1 with AMFR decreases the mean liposome diameter in TEM of freeze-fractured liposomes by about 20%. Incubations without ATP served as control (2 experiments with n = 393, 346 and 223 vesicles analysed (left to right); two-sided Kruskal–Wallis test with Dunn’s post-hoc test: Trx–His–ARL6IP1 and ATP–Trx–His–ARL6IP1 + ATP, P = 0.0024; Trx–His–ARL6IP1 – ATP and control, P < 0.0001; Trx–His–ARL6IP1 + ATP and control, P < 0.0001). Quantitative data shown as the mean ± s.e.m.