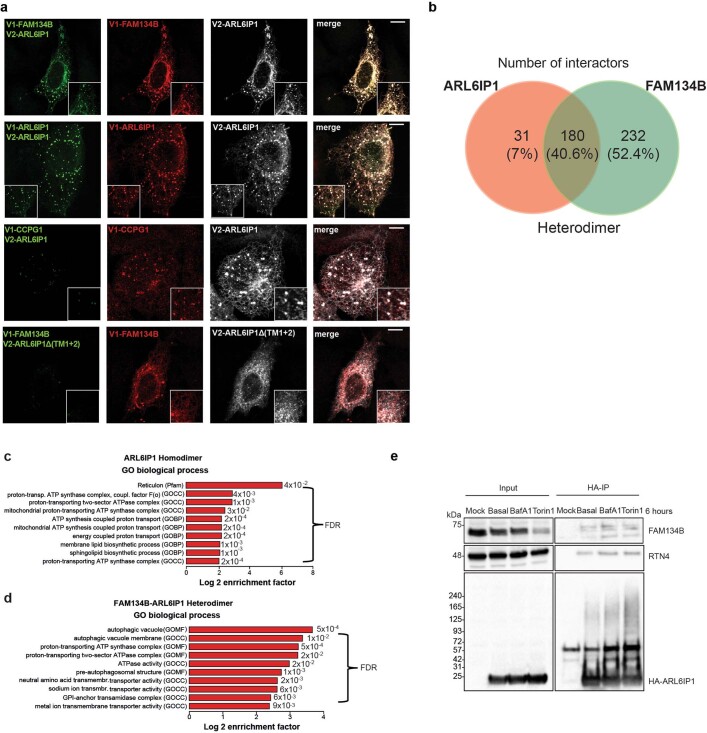
Extended Data Fig. 3. ARL6IP1 forms heterodimers with FAM134B but does not bind LC3B on its own.
a) V1-ARL6IP1/V2-ARL6IP1 homodimers and V1-FAM134B/V2-ARL6IP1 heterodimers are formed in transiently expressing U2OS cells as evidenced by the Venus fluorescence. Co-transfection of V2-ARL6IP1 with V1-CCPG1, another ER resident protein, which does not interact with ARL6IP1, or a construct encoding an ARL6IP1 variant devoid of the first helical hairpin (V2-ARL6IP1Δ(TM1+2)), which is required for the interaction between FAM134B and ARL6IP1, does not result in heteromerisation as evidenced by the lack of the Venus fluorescence. The single channel images represent immunostaining for either FAM134B, ARL6IP1, and CCPG1 or the Venus signal. 1 experiment. Scale bars: 10 µm. b) Venn diagram of interactors of ARL6IP1 and FAM134B homo- and heterodimers, respectively. Numbers represent the identified peptides significantly enriched in three IP and mass spectrometry replicates for each condition. c,d) Annotation enrichment analysis of the interactome of V1-ARL6IP1/V2-ARL6IP1 homodimers and V1-FAM134B/V2-ARL6IP1 heterodimers. Bars represent the significantly enriched gene ontology biological process (GOBP), the gene ontology cellular components (GOCC), the gene ontology molecular function (GOMF), and the domain enrichment (Pfam). The Benjamini-Hochberg FDR value is included (right side of the bars). e) Western blot analysis showing that RTN4, another ER-shaping protein characterised by the presence of a RHD, and FAM134B co-precipitate with HA-tagged ARL6IP1 (1 exp.).