Figure 4. Summary of ploidy, heterozygosity, and genome size diversity in P. parvum.
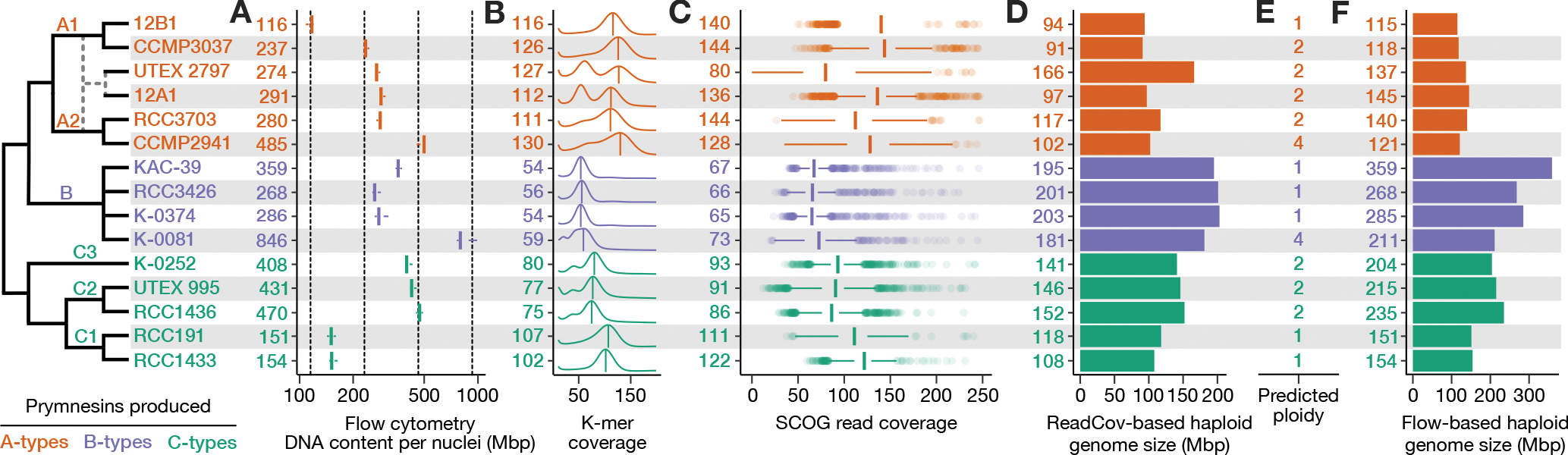
Evolutionary model (left) depicts strain relationships, including a predicted hybridization (dashed lines) giving rise to strains UTEX 2797 and 12A1. A) Boxplots depicting total DNA content per nuclei (Mbp) based on flow cytometry. Numbers (left) indicate mean Mbp per strain. Vertical dashed lines indicate 1x, 2x, 4x, 8x Mbp relative to 12B1. B) K-mer coverage plots depict estimated heterozygosity; numbers indicate CMUKs, i.e., the homozygous k-mer peaks labeled by the vertical bars. Heterozygosity can be qualitatively assessed by the presence and relative height of a second peak at half the k-mer coverage of the homozygous peak. C) Boxplots depicting the distribution of read coverage for 2699 SCOGs; numbers indicate median read coverage. D) Haploid genome size was estimated using read coverage (total read length divided by median Illumina read coverage of SCOGs). E) Predicted ploidy was determined by cross-referencing DNA content (A) with sequencing-based estimated of genome size (B-D). F) Haploid genome size was estimated using flow cytometry (total DNA content divided by proposed ploidy). See also Table S1.
