Figure 1.
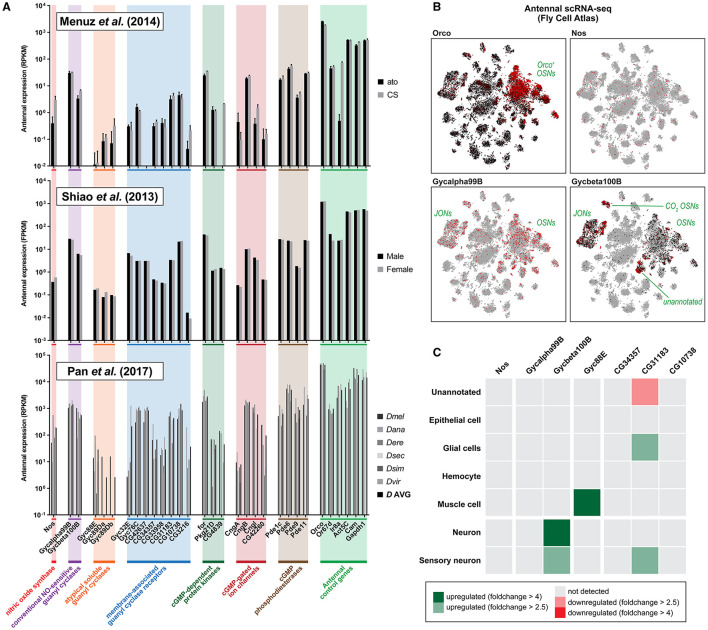
Antenna tissue-specific and antennal single-cell transcriptomics reveal neuronal expression of genes involved in the nitric oxide signaling pathway. (A) Expression of nitric oxide signaling pathway genes across three independent RNA-seq studies sampling antennal tissue only, showing abundant antennal expression of Drosophila nitric oxide synthase (Nos) and both conventional nitric oxide-sensitive guanyl cyclases (Gycα99B, Gycβ100B). Top panel: antennal single-end RNA sequencing of wildtype Canton-S (CS) and atonal (ato) mutant fly antennae, plotting normalized transcript expression in reads per kilobase million (RPKM) (Menuz et al., 2014). Both genotypes retain similar expression levels; notably, Nos is significantly but not entirely depleted in ato mutants which lack coeloconic but retain trichoid and basiconic sensilla. This suggests that the ionotropic receptor (IR) olfactory subsystem also expresses Nos. Middle panel: paired-end RNA sequencing of male and female flies, normalizing expression by fragments per kilobase million (FPKM) (Shiao et al., 2013). Both sexes display comparable expressions. Bottom panel: single-end RNA sequencing of six distinct species plotting normalized transcript expression in RPKM (Pan et al., 2017). A mean average across six species is also plotted (D AVG). Taken together, it is evident that a variety of fruit fly species exhibit comparable gene expression profiles within their antennae. (B) t-distributed stochastic neighbor embedding (tSNE) plots of antennal cells of the Fly Cell Atlas (Li et al., 2022) colored by gene expression (min-max) for four representative genes: Orco, Nos, Gycα99B, and Gycβ100B. Nitric oxide signaling genes show broad expression in Orco+ cells, which label Orco+ OSNs, the olfactory receptor (OR) subsystem. (C) Differential expression (DE) analysis of the above scRNA-seq dataset for some representative genes, by cell-type class compared to the complementary set of all other antennal cells. The nitric oxide-sensitive guanyl cyclase Gycβ100B is differentially expressed in antennal neurons; statistical testing and parametrization are outlined in Supplementary Figures 2, 3 and the Section 2. Colors indicate that statistically positive (green), negative (red), or insignificant (gray) differential expressions are detected at different fold-change thresholds (>4, dark; >2.5, light).