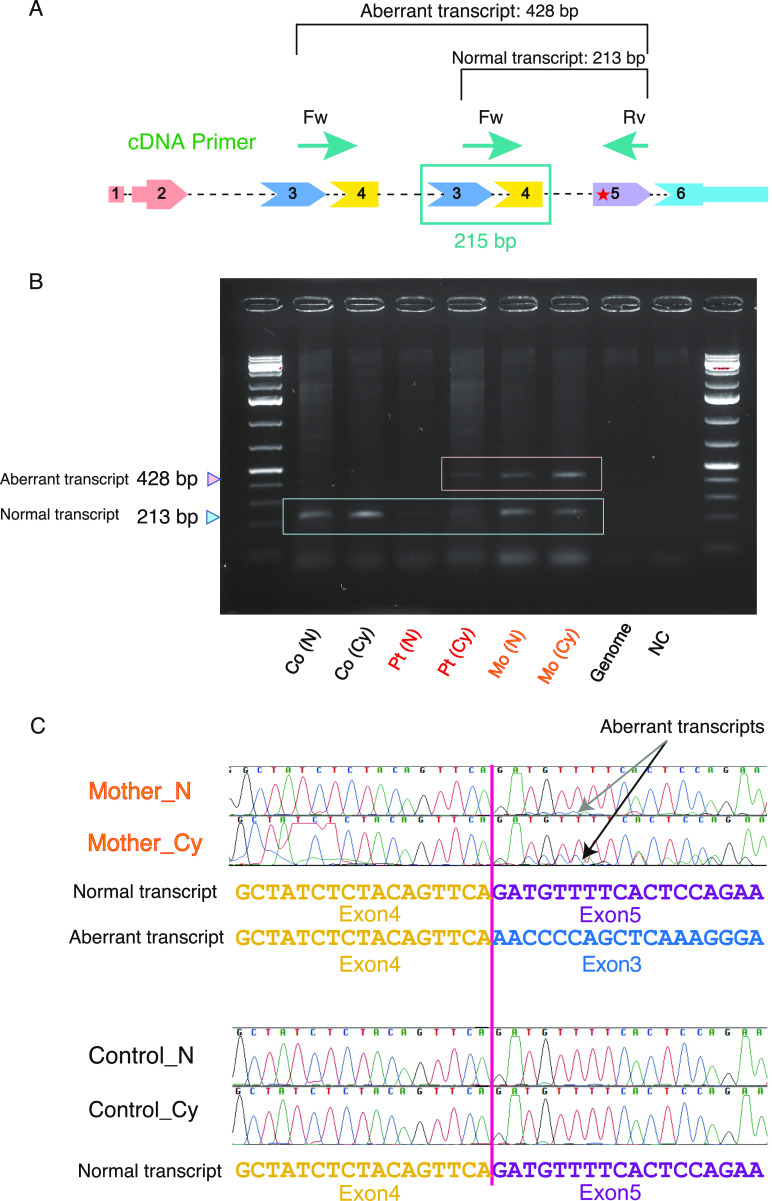
Figure 5. Assessment of NMD of the tandem-duplication allele in patient 1 and her mother by Sanger sequencing.
(A) Schematic presentation of a tandem duplication of exons 3 and 4 in FGF12 of patient 1 and her mother. The predicted splicing pattern is shown by lines connecting the exons. As the duplicated segment included exons 3 and 4, of 215 bp in length, the normal and aberrant transcripts amplified by PCR were estimated to be 213 and 428 bases, respectively. A red asterisk in exon 5 indicates premature termination codons created by the duplication. The positions of primers used in this assay are drawn in green arrows. cDNA, complementary DNA; Fw, forward primer; Rv, reverse primer. (B) Gel electrophoresis of PCR products amplifying normal or aberrant transcripts using cDNA derived from LCLs of patient 1 and her mother. The smaller (213 bp) and larger (428 bp) bands indicate normal and aberrant transcripts, respectively. Co, control; Pt, patient 1; Mo, mother of patient 1; N, treated without CHX (no treatment); Cy, treated with CHX; NC, negative control (no cDNA). (C) Confirmation of NMD in cDNA from a tandem duplication allele. The cDNA sequences of the PCR products showed that cDNA derived from the tandem duplication allele from the mother of patient 1 treated with CHX had higher peaks (black arrow) than that without CHX treatment (gray arrow). No such changes were detected in a control. The primers used in this assay are listed in Table S3 and described in Fig 5A.
Source data are available for this figure.