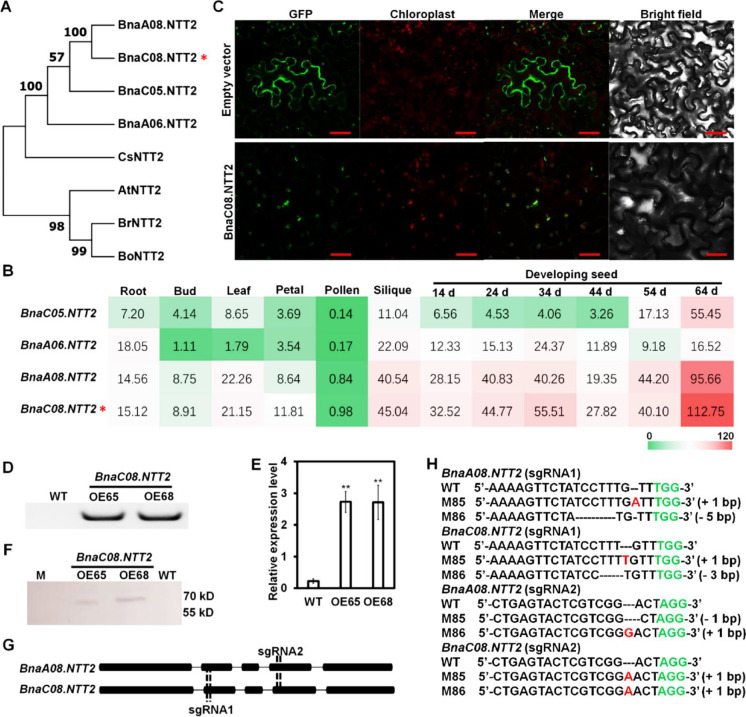
Fig. 1.
Homologous gene analysis of NTT2 and confirmation of BnaNTT2 transgenic plants. A Comparison of amino acid sequence of NTT2 from different plant species using MEGA7. At, Arabidopsis thaliana; Bn, B. napus; Bo, B. oleracea; Br, B. rapa; Cs, Camelina sativa. B The expression of NTT2s in different tissues of B. napus. Gene expression data are from BnTIR (http://yanglab.hzau.edu.cn/). BnaC06.NTT2 is marked in red star in A and B. C BnaC08.NTT2 is localized on chloroplast membrane observed in tobacco epidermal cells under confocal microscopy. Bars = 25 μm. D Identification of the BnaC08.NTT2 overexpression lines using PCR. E Expression level of BnaC08.NTT2 in overexpression lines by quantitative real-time PCR. Total RNA was extracted from leaves of 5-week-old plants. β-actin was used as an internal standard and for normalization. Values are means ± SD (n = 3). **Indicates P < 0.01, based on a student t-test. F Detection of the BnaC08.NTT2 expression in overexpression lines by Western-blot. G CRISPR target sites on NTT2s of B. napus. H Confirmation of BnaC08.NTT2 and BnaA08.NTT2 double mutants (M85 and M86) by sequencing. Insertions are indicated in red and deletions are indicated in black dotted line. PAM sites are indicated in green