Figure 3. Connection between an epigenetic landscape and variable phenotypic outcomes.
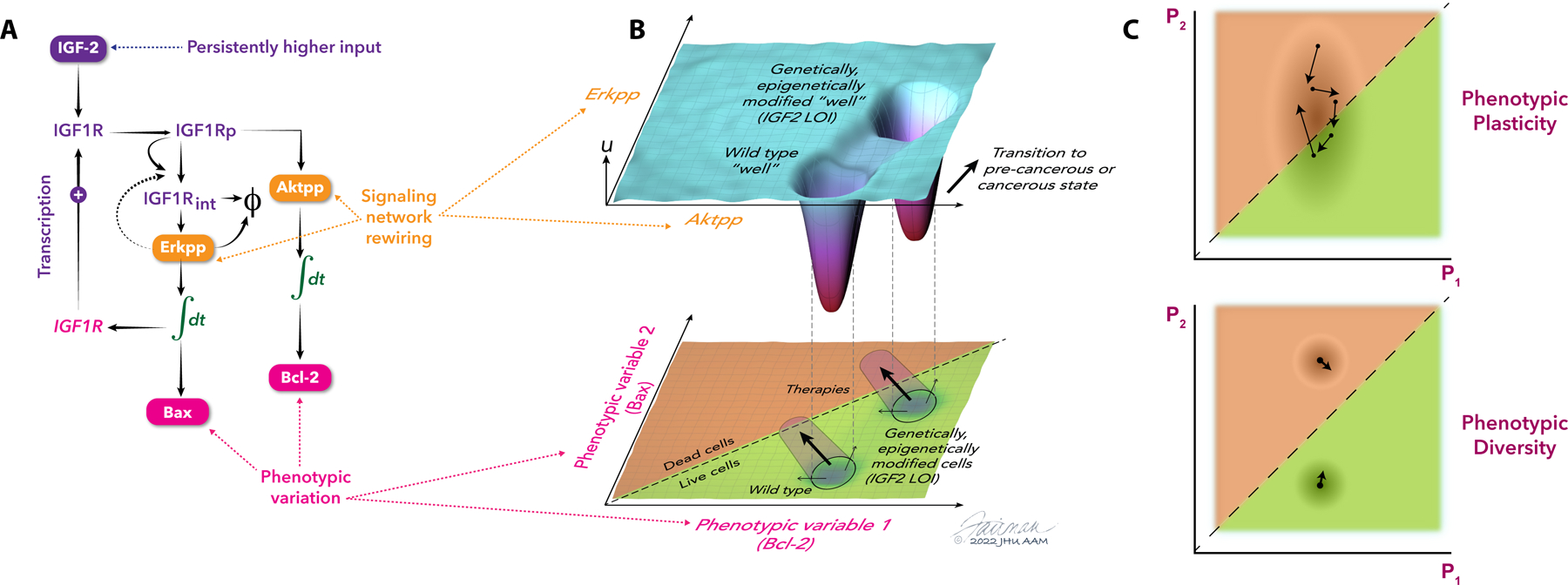
(A) An epigenetic alteration – loss of imprinting (LOI) of the insulin-like growth factor 2 gene (IGF2), implicated in Wilms tumor, doubling of the signaling input, can lead to rewiring of the signaling network activated by the IGF2 receptor IGF1R (depicted as IGF1Rp) through altered receptor trafficking (IGF1Rint), degradation (ϕ) and altered balance of activation of the downstream signaling pathways activating Erk (Erkpp) and Akt (Aktpp) kinases. Rebalancing of Erk and Akt activities translates into transcriptional upregulation of IGF1R, a higher proliferation rate but also rebalancing of pro- and anti-apoptotic protein abundances (BAX versus Bcl-2, respectively) leading to an increased propensity for cell death (113). The integral signs represent integration over time of signaling activities. (B) The landscape alterations that correspond to a change in phenotype (upper panel) is the altered expression and activity of signaling pathway molecules (and thus gene regulatory landscapes in the lower panel) in response to alteration of epigenetic landscapes (IGF2 loss of imprinting, LOI). This leads to emergence of a new attractor in addition to the wild type attractor, resulting in a mosaic wild type/LOI cell distribution in the tissue. This landscape alteration can be ‘mapped’ onto, for example, the apoptosis phenotype-defining network by a quantitative analysis of the dependence of the BCL family protein distributions on the signaling inputs, thus enabling a direct translation of the landscape alterations into phenotype distributions. In this example, the mapping can be visualized as wild type and LOI cell distributions mapped with respect to the areas of cell survival and death on the (BAX, Bcl-2) phenotypic plane, suggesting how treatments targeting LOI cells may be developed to spare the wild type cells. Arrows in the lower panel represent the effect of drugs, such as IGF1R inhibitors shifting the landscape and phenotypic distribution towards the boundary separating survival and death, with the red areas depicting the effect on the wild type and LOI cell populations. (C) A more general view of landscape mapping onto the apoptosis phenotypic plane. By analogy with Fig. 1B, one can contrast mapping of a large attractor versus two more limited attractors, representing the difference between a plastic and stochastic state (phenotypic plasticity) versus a state with two alternative stable attractors (phenotypic diversity). The more plastic state can allow cells to escape from the death area to the survival area even in the presence of a treatment (such as in (B)), by stochastically ‘exploring’ the available attractor, whereas a combination of more stable attractors (with the same overall entropy as the more plastic state) can allow for selective targeting by one but not the other attractor. Therefore, the treatment strategy suggested in (B) may benefit from the initial intervention stabilizing smaller attractors within a larger one and thus decreasing the plasticity of the state, particularly through epigenetic perturbations.
