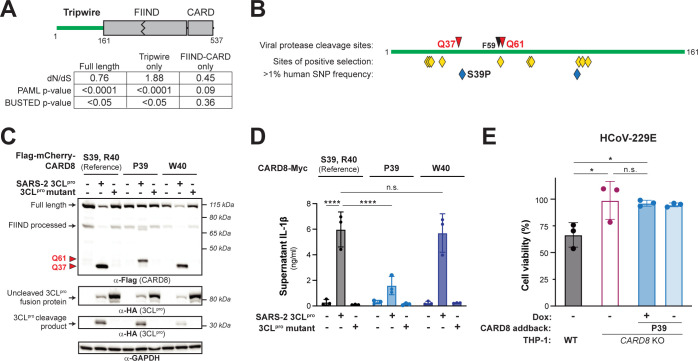
Fig 4. Human polymorphism in CARD8 reduces sensing of coronavirus 3CLpro.
(A) Evolutionary analyses of positive selection were performed on full length CARD8 (encoding residues 1–537), the disordered N-terminal “tripwire” region (encoding residues 1–161), and the FIIND-CARD region (encoding residues 162–537). P values from PAML and BUSTED analyses are shown, along with the dN/dS value obtained from PAML. (B) Schematic of the CARD8 “tripwire” region. Red and black triangles and amino acid numbers indicate sites of 3CLpro and HIV-1pro cleavage, respectively. Yellow diamonds indicate codons predicted to be evolving under positive selection by at least one evolutionary analysis (S4 Table). Blue diamonds indicate high frequency (>1% allele frequency) nonsynonymous SNPs in humans (S5 and S6 Tables). The position of the S39P substitution that results from SNP rs12463023 is shown. (C, D) Reference human CARD8 (S39, R40) or human CARD8 variants (P39 or W40) were coexpressed with the indicated protease construct and assayed for 3CLpro-mediated cleavage (C) or CARD8 inflammasome activation (D). (E) The CARD8 variant P39 was complemented into CARD8 KO1 THP-1 cells using a Dox-inducible lentiviral construct and were infected along with WT and CARD8 KO1 THP-1 cells with hCoV-229E in the presence or absence of 100 ng/mL Dox. Cell viability was measured using the Cell Titer Glo assay, 48 h post-infection. Individual values (n = 3), averages, and standard deviations shown are representative of experiments performed at least twice. Data were analyzed using two-way ANOVA with Šidák’s post-test (D) or one-way ANOVA with Tukey’s post-test (E). * = p < 0.05, **** = p < 0.0001, n.s. = not significant. Data for Fig 4D and 4E can be found in S1 Data. Dox, doxycycline; SNP, single nucleotide polymorphism; WT, wild-type; 3CLpro, 3CL protease.