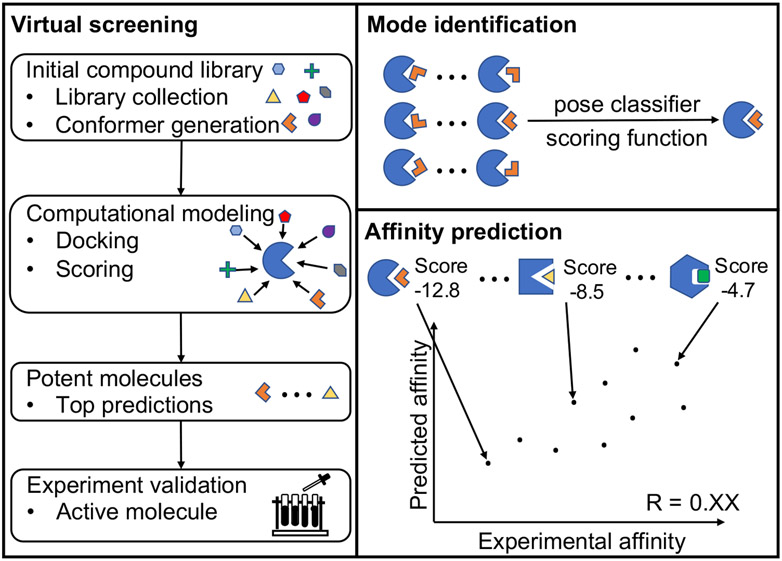
Figure 1:
Three major applications of an RNA-ligand interaction model. Virtual screening involves docking against, small molecules in a large library and scoring every docked pose. Top-scored selections are treated as the most, promising candidates for putative binders. For a given RNA-ligand pair, computational models for ligand binding pose identification and RNA-ligand binding affinity prediction rely on scoring the possible RNA-ligand complex structures. An ideal scoring function for ligand binding pose identification should have the ability to distinguish the native pose from a large pool of docked decoy poses, while achieving the maximum correlation between the predicted scores and the experimental affinities for different. RNA-ligand pairs.