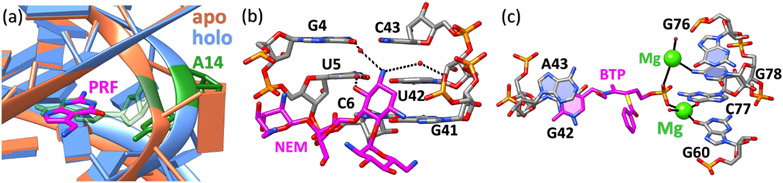
Figure 2:
RNA conformational changes and binding interactions mediated by water molecules and ions. (a) The local structure difference of preQ1 riboswitches between apo (ligand-free) and holo (ligand-bound) states. The structure in orange denotes the apo state (PDB code: 6VUH (113)) and the structure in blue denotes the holo state (PDB code: 3Q50 (114)) with its bound small molecule (PRF) colored in magenta. Upon binding, the small molecule displaces residue A14 (colored in green for both apo and holo states) and causes the local structural transition. (b) Water molecules mediated RNA-ligand interactions. Water molecules form a bridge between small molecule Neomycin B (NEM, magenta) and 16S-rRNA A-site (PDB code: 2ET4 (115)). The isolated red dots denote the oxygen atoms in water molecules. The black dashed lines show the water-mediated hydrogen bonding contacts that promote NEM binding to the RNA receptor. (c) Metal ions in RNA-small molecule interactions. The ligand benfotiamine (BTP, magenta) interacts with residues G60, C77, and G78 of the Thi-box riboswitch through two magnesium ions (green) and the G42-A43 base stack (PDB code: 2HOO (116)). The black solid lines represent the inner sphere metal ion coordination. The polyanionic RNA recognizes the positively charged metal ion complex made up of the monophosphorylated compound and cations.