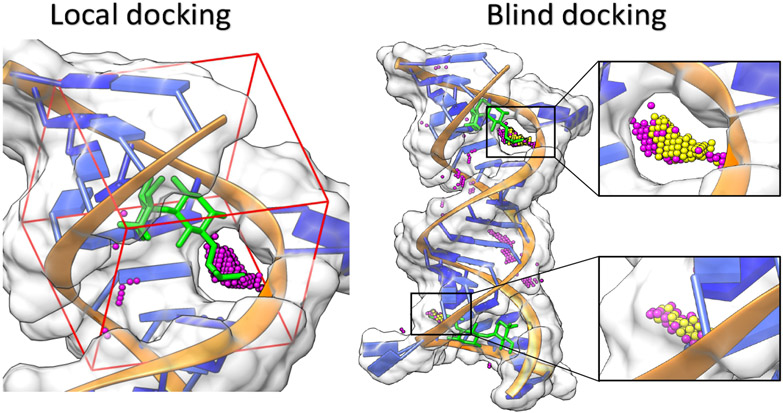
Figure 3:
The difference between local and blind docking. A complex of an aminoglycoside antibiotic, gentamicin (green) and the 16S-rRNA A site of bacterial ribosome is used for illustration (PDB code: 2ET3 (115)). In this example, both docking (local & blind) processes are carried out using the RLDOCK model (70; 71). In local docking, the binding pocket is predefined and the sampling is contained within the red dashed box. The small magenta spheres denote candidate binding sites predicted by RLDOCK. In blind docking, the binding site detection is performed across the whole surface of the RNA. The small yellow and magenta spheres denote the predicted high- and low-probability binding sites, respectively. Two cavities identified by RLDOCK (anchored by yellow spheres) are zoomed out separately.