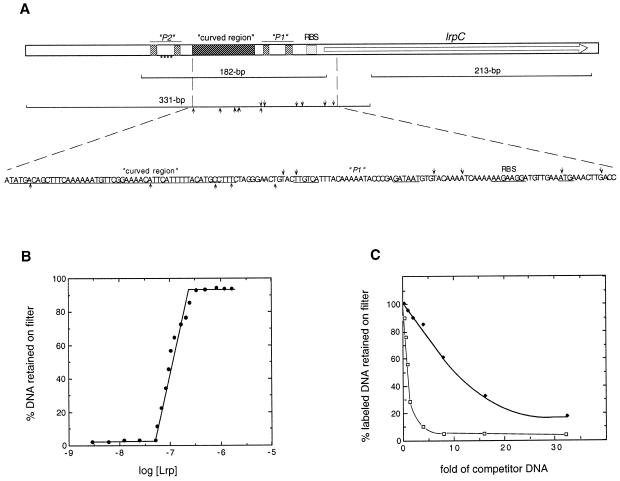
Figure 2.
LrpC binds preferentially to curved DNA. (A) Schematic representation of the DNA fragments used in this work. The lrpC gene is indicated with an arrow. Putative promoters P1 and P2 (pale grey box), the computer predicted curved region (dark grey box) and the ribosomal binding site (RBS) are indicated. A nucleotide sequence resembling the consensus EcoLrp binding site, within promoter P2, is indicated. The DNA fragments used in this work are shown. The relevant features of the nucleotide sequence that shows DNase I hypersensitive sites are indicated. The arrows denote DNase I hypersensitive sites in the upper and lower strand, respectively. (B). The 331 bp curved [γ-32P]DNA fragment (1 nM) was incubated with increasing LrpC concentrations (2–1200 nM) in buffer B containing 75 mM NaCl for 15 min at 30°C. The DNA retained on the filter was corrected for the retention of [32P]DNA in the absence of LrpC protein (2–3% of total input). (C) Competition assays of LrpC protein DNA binding activity. The 331 bp curved [γ-32P]DNA fragment (1 nM) was incubated with LrpC (100 nM) and the indicated fold excess of cold 331 bp curved DNA fragment (square) or cold 213 bp non-curved DNA fragment (diamond). Binding conditions and determination of retained LrpC–DNA were as in (B). Under these conditions, in the absence of competitor DNA, 60% of the labelled DNA was retained, and this value is given as 100%.