Fig. 3. Conserved function of constrained transcription factor binding sites.
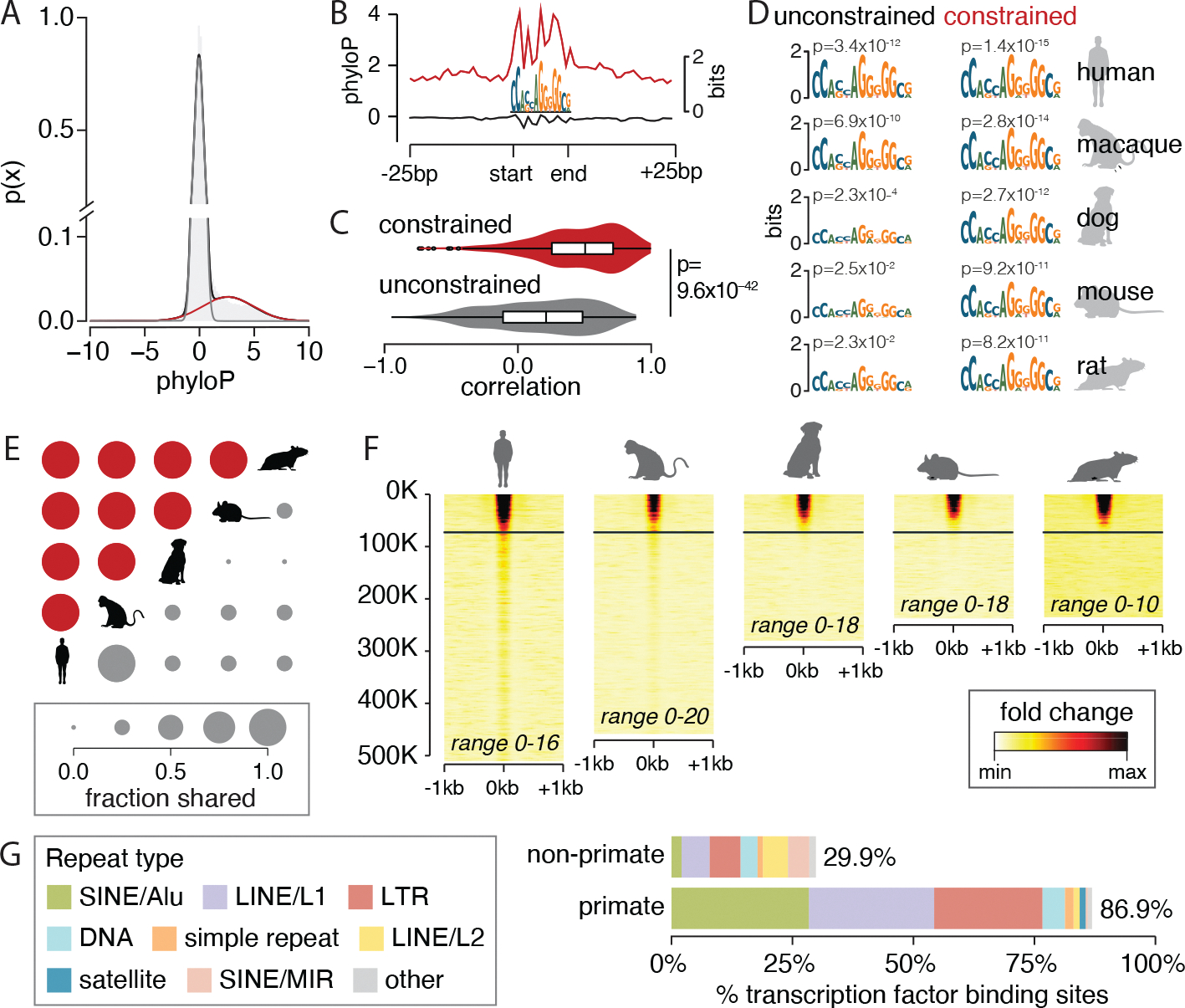
(A) A two-component Gaussian mixture model fit over average phyloP scores across binding sites for CTCF distinguishes the distribution for evolutionarily constrained sites (red) from others (gray). (B) At CTCF binding sites, aggregate phyloP scores are high for constrained binding sites (red, 61,832 sites) but not for unconstrained binding sites (gray, 424,177 sites). The same pattern is observed for other transcription factors (fig. S10). (C) Across all transcription factors, aggregate phyloP scores are more strongly correlated (Pearson’s correlation) with binding site information content for constrained sites than for unconstrained sites. Boxes and whiskers represent 25% quartile, 75% quartile, minimum, and maximum, with a horizontal line at the median. The shading indicates the density of the data. (D) CTCF logos of constrained and unconstrained sets for four species made by lifting over human transcription factor binding sites. (E) Fraction of constrained (red) and unconstrained (gray) CTCF binding sites that are shared between pairs of species. (F) CTCF transcription factor chromatin immunoprecipitation sequencing (ChiP-seq) signal over binding sites in mammalian livers sorted by average phyloP scores. Each row is a binding site; in nonhuman species, only aligned sites are shown. The horizontal lines indicate significant constraint. Ranges give the minimum and maximum ChIP-seq fold change over input for each species. (G) Percentage of primate-specific and non–primate-specific transcription factor binding sites that are derived from individual transposable element classes. LINE, long interspersed nuclear element; LTR, long terminal repeat; MIR, mammalian-wide interspersed repeat; SINE, short interspersed nuclear element. [Species silhouettes are from PhyloPic]
