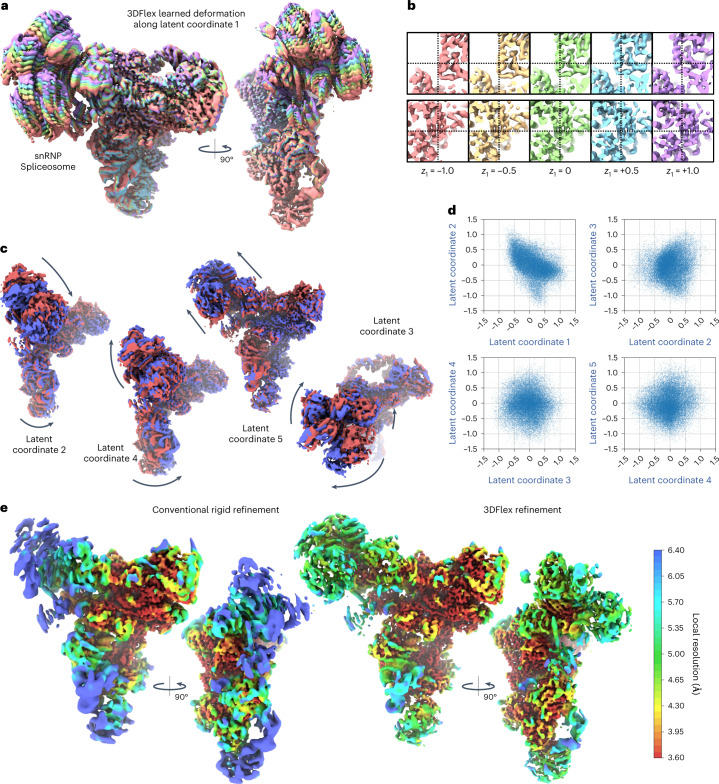
Fig. 2. 3DFlex with a 5D latent space applied to particle images of an snRNP Spliceosome complex, demonstrating the capacity for 3DFlex to resolve multiple modes of nonrigid deformation simultaneously, while also capturing high-resolution structural detail.
See Supplementary Video 1. a, Colored series of convected densities learned by 3DFlex, at five positions along the first latent dimension, ranging from minus one to plus one standard deviation. b, Same as a but focused on key structural details in the head region of the protein. The top row shows an α-helix that translates several Angstroms. The bottom row shows a β-sheet, which translates and deforms. c, Convected densities from 3DFlex at minus one (red) and plus one (blue) standard deviations in the latent space, along each of the remaining four latent dimensions. Each dimension resolves a different type of motion within the same model. d, Scatter plots showing the final distribution of learned particle latent coordinates across the dataset. e, The left shows a density map from conventional nonuniform rigid refinement colored by local resolution. The right shows a canonical density map from 3DFlex, colored on the same local resolution color scale. The two maps are filtered by local resolution to aid in visualizing weak density in low-resolution areas in the conventional refinement.