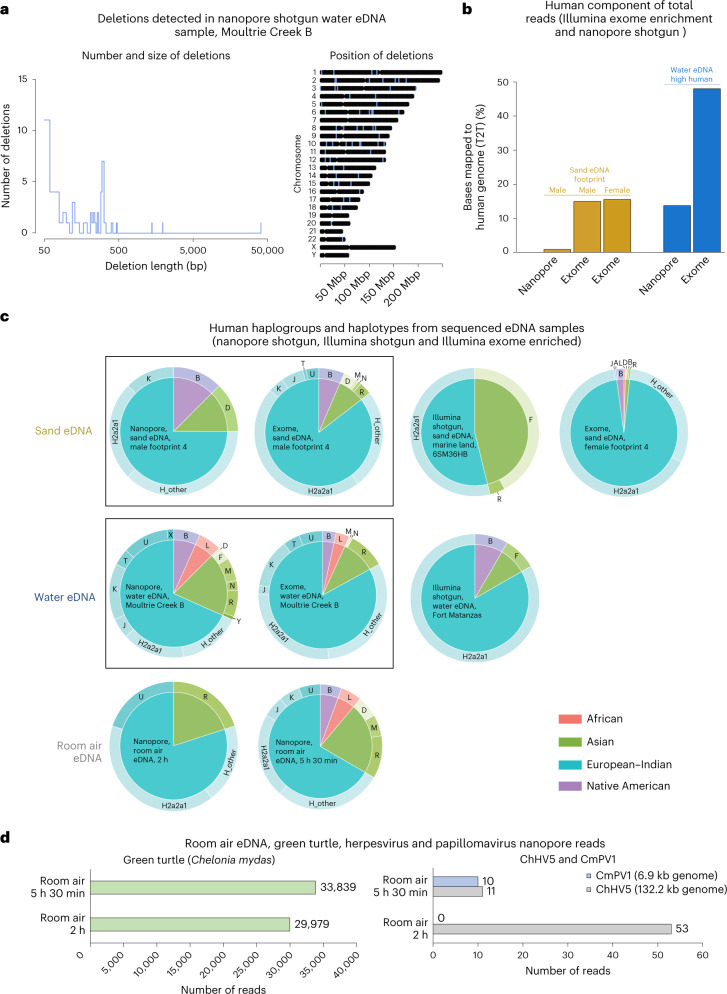
Fig. 4. Mutation and genetic ancestry analysis of shotgun and exome-enriched eDNA samples.
a, Known human genomic deletions (gnomAD database) detected in a nanopore shotgun water sample, including the number and size of deletions (left) and the genomic position of each deletion (right); deletion locations are denoted by blue shading. The full details of the detected deletions can be found in Supplementary Table 5. b, Percentage of sequenced reads from eDNA metagenomic samples that aligned (minimap2) to the human genome, compared with all reads generated for each sample (regardless of species of origin). Nanopore shotgun sequencing is compared with Illumina exome enrichment. Note that the same male footprint sand eDNA sample and the same high-human water eDNA sample was used for both shotgun sequencing (nanopore) and exome enrichment (Illumina). c, Haplogroup and haplotype analysis of human-mitochondrial-aligning reads for each sequenced (shotgun or exome-enriched) intentional human sample and for the water and sand bycatch Illumina shotgun-sequenced samples (sea turtle focused) with the highest number of human-aligning reads. Pie charts within the same box are from sequencing data for which the same original sample was used (that is, for shotgun and exome enrichment). d, Quantification of green sea turtle and sea turtle herpesvirus (ChHV5) and papillomavirus (CmPV1) aligning reads (minimap2) from the same room-air eDNA nanopore shotgun sequencing data as were used for identifying human-aligning reads (Fig. 3a).