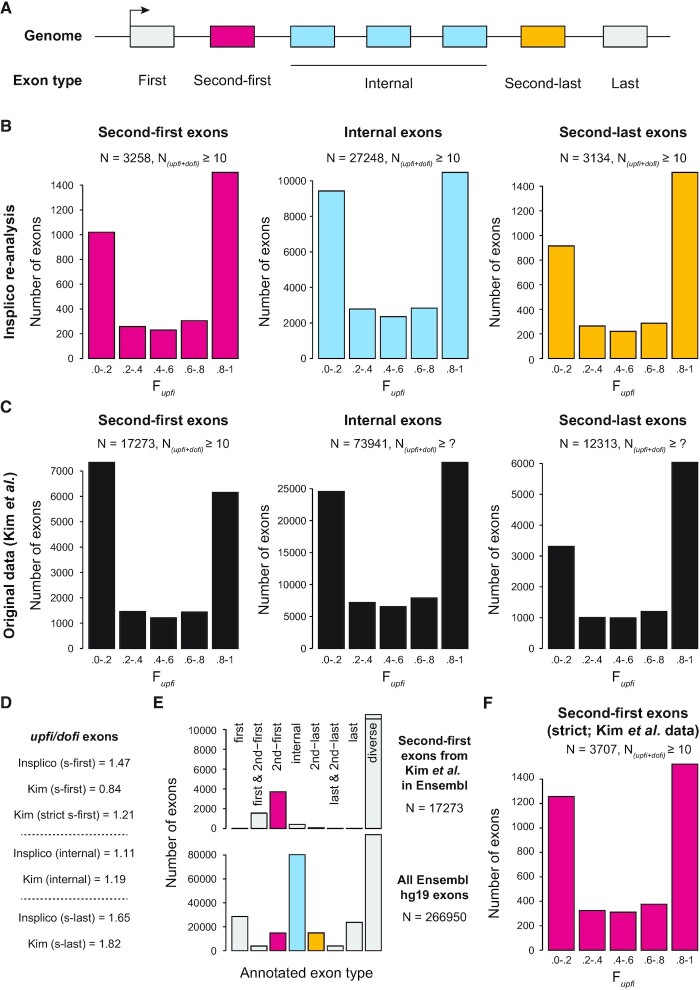
Figure 3.
Reproduction of major AISO patterns from Kim et al. (2017) using Insplico. (A) Schematic representation of the different groups of exons investigated for a representative gene. (B) Distribution of exons according to Fupfi values generated by Insplico for second-first (left), truly internal (middle) and second-last (right) exons as defined by extract_exons_from_gtf.pl. (C) Distribution of exons according to Fupfi values for second-first (left), truly internal (middle) and second-last (right) exons as provided by Kim et al. N(upfi + dofi) for internal and second-last exons was not provided in the original publication. (D) Distribution of second-first exons as annotated by Kim et al. according to exon types extracted by extract_exons_from_gtf.pl. ‘Diverse’ correspond to exons with more than one type of annotation across transcripts. (E) Distribution of exon types as extracted by extract_exons_from_gtf.pl for all exons in the Ensembl hg19 annotation. (F) Distribution of strictly defined second-first exons from Kim et al.’s original set according to Fupfi values as provided by Kim et al. (15).