Abstract
Facioscapulohumeral muscular dystrophy (FSHD) is one of the most prevalent neuromuscular disorders. The disease is linked to copy number reduction and/or epigenetic alterations of the D4Z4 macrosatellite on chromosome 4q35 and associated with aberrant gain of expression of the transcription factor DUX4, which triggers a pro-apoptotic transcriptional program leading to muscle wasting. As today, no cure or therapeutic option is available to FSHD patients. Given its centrality in FSHD, blocking DUX4 expression with small molecule drugs is an attractive option. We previously showed that the long non protein-coding RNA DBE-T is required for aberrant DUX4 expression in FSHD. Using affinity purification followed by proteomics, here we identified the chromatin remodeling protein WDR5 as a novel DBE-T interactor and a key player required for the biological activity of the lncRNA. We found that WDR5 is required for the expression of DUX4 and its targets in primary FSHD muscle cells. Moreover, targeting WDR5 rescues both cell viability and myogenic differentiation of FSHD patient cells. Notably, comparable results were obtained by pharmacological inhibition of WDR5. Importantly, WDR5 targeting was safe to healthy donor muscle cells. Our results support a pivotal role of WDR5 in the activation of DUX4 expression identifying a druggable target for an innovative therapeutic approach for FSHD.
INTRODUCTION
Facioscapulohumeral muscular dystrophy (FSHD) is an inherited progressive neuromuscular disorder and afflicts both children and adults regardless of gender. At onset, FSHD patients usually present with weakness of the muscles of the face (facio), shoulder (scapulo) and upper arms (humeral). Disease progression is usually slow and might involve additional muscle groups. FSHD patients frequently display asymmetric distribution of muscle damage and are characterized by high variability in age of onset, rate of progression and disease severity, even in family relatives (1). Notably, several monozygotic-twin discordances for FSHD have been reported (2–6). Although the molecular basis of this heterogeneity is not fully understood, an increasing body of evidence suggests that it derives from the interplay of complex genetic and epigenetic events (7).
FSHD is associated to copy number reduction and/or epigenetic alterations of the D4Z4 macrosatellite repeat located on chromosome 4q35 (8–11). In healthy subjects, the FSHD locus displays features of repressed chromatin such as relatively high levels of DNA methylation or trimethylation of histone H3 lysine 9 or 27, and is poorly transcribed. In FSHD patients, a local and partial loss of repressive features facilitates transcription of the FSHD locus (12) leading to the aberrant expression of the double homeobox 4 (DUX4) gene in a subset of FSHD muscle nuclei (13).
DUX4 encodes for a transcription factor physiologically involved in zygotic genome activation during the cleavage stage of early embryonic development (14–16). DUX4 is epigenetically silenced in most adult somatic tissues, including skeletal muscle. Aberrant reactivation of DUX4 expression in FSHD triggers several pathways toxic to skeletal muscle, including inhibition of myogenic differentiation and cell death (reviewed in (1)). Consequently, treatments reducing DUX4 levels in FSHD muscle cells would have high therapeutic relevance.
While we and others have identified many factors responsible for maintaining DUX4 repressed in healthy subjects (9–11,17–22), relatively little is known regarding factors required for DUX4 expression in FSHD muscle cells (19,23).
We previously identified the long non-protein coding RNA (lncRNA) D4Z4 binding element-transcript (DBE-T) (19). Intriguingly, global DNA methylation analysis identified DBE-T as the top most hypomethylated region in FSHD also showing that it is less methylated in more affected compared to less affected muscles (24). DBE-T is a chromatin-associated lncRNA working in cis on the FSHD locus and required for the expression of DUX4 (19). We previously showed that the histone methyltransferase ASH1L mediates, at least in part, DBE-T activity at the FSHD locus (19). Nevertheless, DBE-T silencing causes a much stronger DUX4 downregulation than ASH1L knockdown (19), suggesting that additional factors might be required for DUX4 activation by DBE-T. To elucidate this aspect, we investigated the molecular mechanism by which DBE-T activates gene expression. Here, we found that the chromatin remodeling factor WD Repeat Domain 5 (WDR5) specifically and directly interacts with DBE-T. WDR5 is recruited to the FSHD locus in a DBE-T dependent manner and activates DUX4 expression in FSHD muscle cells. Accordingly, WDR5 knockdown or pharmacological inhibition rescue cell viability and myogenic differentiation of FSHD muscle cells without affecting healthy muscle cells. Our results reveal a novel DUX4 activator and open new perspectives for the treatment of FSHD patients.
MATERIALS AND METHODS
Cell culture
CHO (ATCC: CCL-61), HeLa (ATCC: CCL2) and HEK293 Flp-In T-REx (Invitrogen, #R78007) cells were grown in DMEM high glucose medium with l-glutamine (EuroClone, #ECB3000) and Sodium Pyruvate (EuroClone, #ECM0542D) supplemented with 10% Fetal Bovine Serum (FBS, Thermo Fisher Scientific, #A4766801) and 1% penicillin/streptomycin (Pen/Strep, Thermo Fisher Scientific, #15140122).
HEK293 Flp-In T-REx BirA-Flag-Csy4 cells were generated according to manufacturer instructions and the expression of the BirA-Flag-Csy4 protein was obtained by the administration of doxycycline for 24 h at the indicated concentration.
Human primary muscle cells from healthy donors and FSHD patients were obtained from the University of Rochester Fields Center for FSHD. Human immortalized FSHD muscle cells were obtained from the University of Massachusetts Medical School Senator Paul D. Wellstone Muscular Dystrophy Cooperative Research Center for FSHD.
Muscle cells were cultured in a humidified atmosphere at 37 °C with 5% O2 and 5% CO2. Primary muscle cells were maintained in proliferating media Ham's F-10 medium (Nutrient Mixture F10 Ham, with l-glutamine and sodium bicarbonate, Sigma, #N9608) supplemented with 20% fetal bovine serum (FBS, Thermo Fisher Scientific, #A4766801), and 1% penicillin/streptomycin (Pen/Strep, Thermo Fisher Scientific, #15140122), 10 ng/ml basic fibroblast growth factor (bFGF, Tebu-bio, #100-18B), and 1 μM dexamethasone (Sigma-Aldrich, #D4902). To induce differentiation, myoblasts were plated at a confluence of 45 000 cells/cm2 and, 24 h after seeding, growth medium was replaced by differentiation media composed by DMEM:F12 (1:1, Sigma-Aldrich, #D9785) supplemented with 20% knockout serum replacement (KOSR, Thermo Fisher Scientific, #10828028), 3.151 g/l glucose, 10 mM MEM non-essential amino acids (Thermo Fisher Scientific, #11140050), 100 mM sodium pyruvate (Thermo Fisher Scientific, #11360070). Differentiation was carried out for 96 h changing the media every other day.
Immortalized muscle cells were cultured in gelatine coated plates and in a humidified atmosphere at 37°C with 5% O2 and 5% CO2. Cells were maintained in growth media DMEM-HIGH Glucose (Sigma-Aldrich, #D5671) and Medium 199 (Sigma-Aldrich, #M2520) in a ratio 4:1, supplemented with 15% FBS (FBS, Thermo Fisher Scientific, #A4766801), 1.4 mg/l vitamin B12 (Sigma-Aldrich, #V6629), 0.03 mg/l ZnSO4 (Sigma-Aldrich, #83265), 0.055 mg/l dexamethasone (Sigma-Aldrich, #D4902), 2.5 ug/l hepatocyte growth factor (HGF,Tebu-bio, #100-39), 10 ug/l fibroblast growth factor (FGF, Tebu-bio, #100–18B), 0.02M 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, Sigma-Aldrich, #54457), 1% Pen/Strep (Thermo Fisher Scientific, #15140122). Immortalized cells were induced to differentiate using differentiation media composed by DMEM-HIGH Glucose (Sigma-Aldrich, #D5671) and Medium 199 (Sigma-Aldrich, #M2520) in a ratio 4:1, supplemented with 2% Donor Horse Serum (EuroClone #ECS0091L) and 1% Pen/Strep (Pen/Strep, Thermo Fisher Scientific, #15140122). Differentiation was carried out for 96 h changing the media every other day.
Constructs and cloning procedures
The construct pHM/GWA Csy4 H29A S50C was a gift from Jennifer Doudna (Addgene plasmid # 45029; http://n2t.net/addgene:45029; RRID:Addgene_45029) (25) and used to amplify and clone, using the gateway strategy (Invitrogen) the sequence of Csy4 in frame into pcDNA/FRT/TO/NLS/BirA/Flag construct, which was gently provided by Dr G. Superti-Furga.
Given that DBE-T contains multiple copies of the same sequence at its 3’-end (19) (Supplementary Figure S1A), for the Gal4-λN-BoxB targeting system we used a non-redundant 5’-end fragment (Supplementary Figure S1A). This region, corresponding to the fragment nt 4317–7463 of AF117653, was amplified and cloned into pGEM-T vector (Promega, #A3600). This portion of DBE-T was then virtually divided in 7 partially overlapping fragments (Figure 1C) of equal size and each fragment was amplified by PCR and cloned into pGEM-T vector. The same cloning strategy was used for a non-coding portion of the LacZ gene, used as negative control. Each insert was sequence verified.
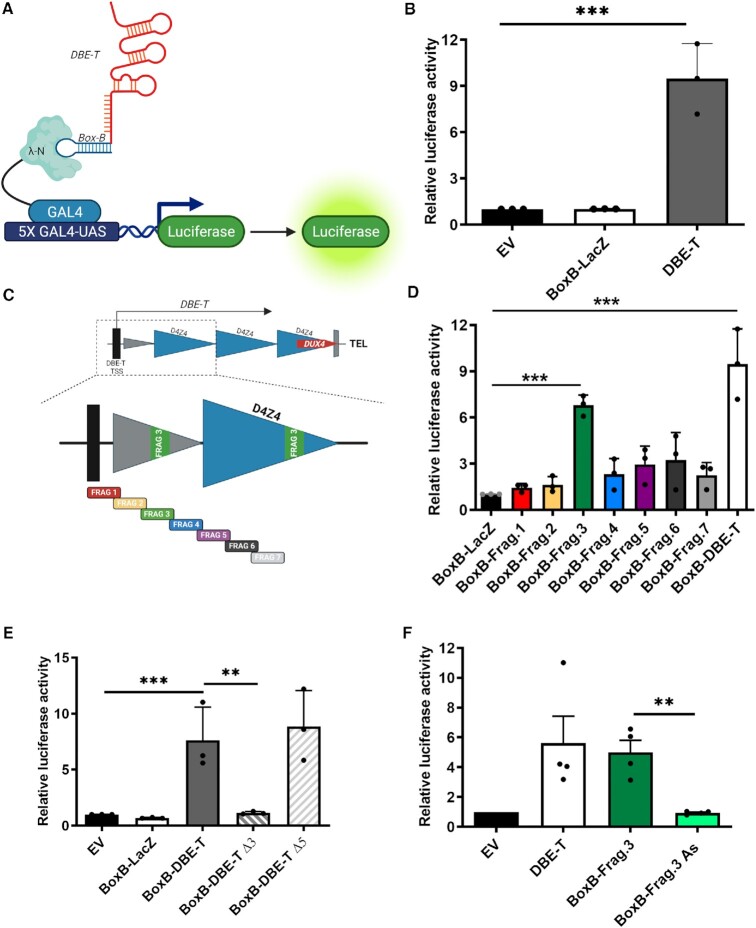
Figure 1.
A reporter system to study DBE-T activity in mammalian cells. (A) Schematic representation (not in scale) of the Gal4-λN-BoxB targeting system to study DBE-T ability to regulate transcription in which the firefly Luciferase reporter gene is under the control of five copies of the sequence recognized by the yeast transcription factor GAL4 (5X GAL4-UAS). The GAL4 DNA binding domain fused to the λN RNA-binding domain that specifically recognizes the BoxB sequence fused with DBE-T is shown. Figure created with BioRender.com. The RNA structure is based only on BioRender templates. (B) Luciferase activity measured in CHO cells transfected with Gal4-λN and 5X GAL4-UAS-Luciferase in combination with empty vector (EV), BoxB-LacZ or BoxB-DBE-T. Luciferase activity relative to EV control is shown. N = 3. Student's t-test. ***P< 0.001. (C) Top. Schematic representation (not in scale) of the FSHD locus (see Supplementary Figure S1A for details). Bottom. Enlargement showing the DBE-T portion and its non-redundant fragments used for the luciferase assays (see Materials and Methods for details). (D) Luciferase activity in CHO cells transfected with the Gal4-λN-BoxB targeting system in combination with BoxB-fused to LacZ, the indicated DBE-T fragments or DBE-T. Luciferase activity relative to BoxB-LacZ control is shown. N = 3 Student's t-test. ***P< 0.001. (E) Luciferase activity in CHO cells transfected with the Gal4-λN-BoxB targeting system in combination with empty vector (EV) or BoxB-fused to LacZ, DBE-T, DBE-T deleted of Frag.3 (Δ3) or DBE-T deleted of Frag.5 (Δ5). Luciferase activity relative to EV control is shown. N = 3. Student's t-test. **P< 0.01, ***P< 0.001. (F) Luciferase activity in CHO cells transfected with the Gal4-λN-BoxB targeting system in combination with empty vector (EV) or BoxB-fused to DBE-T, DBE-T Frag.3 or the antisense of DBE-T Frag.3 (AS). Luciferase activity relative to EV control is shown. N = 3. Student's t-test. **P< 0.01.
For the generation of BoxB-tagged RNAs, the DBE-T fragments inserted in pGEM-T were excise and cloned into the BoxB pcDNA 3.0 vector, by NotI (New England BioLabs, #R0189) and BamHI (New England BioLabs, #R0136) restriction enzymes. BoxB in pcDNA3 was a gift from Howard Chang (Addgene plasmid # 29727; http://n2t.net/addgene:29727; RRID:Addgene_29727) (26).
The DBE-T mutants deleted of Frag.3 or Frag.5 were respectively generated by removing Frag.3 with the restriction enzymes SfiI and KpnI, while Frag.5 with BlpI, followed by DNA ligation.
For the generation of Csy4-tagged RNAs, three copies of the Csy4 RNA hairpin were initially cloned into pcDNA 3.0 using EcoRI (New England BioLabs, #R0101) and BamHI restriction enzymes. The sequences of DBE-T Frag.3 or its antisense were then cloned downstream of the Csy4 tag through EcoRV (New England BioLabs, #R0195) restriction enzyme.
The sequence of human WDR5 cloned into the pDonor221 was gently provided by Dr G. Superti-Furga and used to clone through the gateway strategy the cDNA into the pGEX-6P-1 (Merck, # GE28-9546-48) and pcDNA-FRT-TO plasmids to produce GST-WDR5 and STREP-HA-WDR5, respectively.
The list of primers used in this study is provided in Supplementary Table S1.
Transfection and transduction
WDR5 and non-silencing control siRNAs were purchased from Dharmacon (SMARTpool: ON-TARGETplus Human WDR5 siRNA, #FE5L013383000005; non-targeting control, #FE5D0018101020) and used at a final concentration of 25 nM. The siRNA for DUX4 was purchased from Thermo Fisher Scientific (Custom Stealth siRNA). Control shNS and shDBE-T shRNAs cloned in pLKO lentiviral vectors were previously described (19).
The list of siRNAs and shRNAs used in this study is provided in Supplementary Table S1.
Transfection of siRNAs was performed by using Lipofectamine 3000 Transfection Reagent (Thermo Fisher Scientific, #L3000001), following the manufacturer's instructions. The cells were seeded at 90% confluence and after 24 h differentiation was induced. The siRNAs were delivered 24 h after induction of myogenic differentiation. Medium was replaced the day after transfection and myotubes were harvested 96h after induction of differentiation.
Plasmids were delivered by using Lipofectamine LTX Reagent with PLUS Reagent (Thermo Fisher Scientific, #A1262), following the manufacturer's instructions.
When transfection of both siRNAs and plasmids was required, cells were reverse-transfected with siRNAs by using Lipofectamine 3000 Transfection Reagent and the day after they were transfected with the respective plasmids by using Lipofectamine LTX Reagent with PLUS Reagent, following manufacturer's instructions. Cells were harvested 48 h after the second transfection.
Production of shNT- and shDBET-pLKO lentiviral particles was carried out as previously described (19). Lentiviral transduction of human immortalized FSHD myoblasts was carried out as previously described (27).
Generation of the flp-in T-REx HEK293 NLS-BirA-flag-csy4 cell line
The pcDNA5/FRT/TO/BirA/FLAG/Csy4 or pcDNA5/FRT/TO/NLS/BirA/FLAG/Csy4 were co-transfected with the pOG44 Flp-Recombinase Expression vector (Thermo Fisher Scientific, #V600520) into Flp-In T-REx HEK293 cells to generate the BirA/FLAG/Csy4 or BirA/FLAG/NLS/Csy4 inducible cell lines, according to the vendor protocol. Cells were then selected using both Blasticidin (Thermo Fisher Scientific, #A1113902) and Hygromycin B (Thermo Fisher Scientific, #10687010), and used as polyclonal population.
RNA extraction, reverse transcription and quantitative real-time PCR
Total RNA was extracted using PureLink RNA Mini Kit (Thermo Fisher Scientific, #12183025), following the manufacturer's instructions. Briefly, cells were lysed in Lysis buffer supplemented with 2-mercaptoethanol and homogenized by passing the lysate 5–10 times through a 21-gauge syringe needle. After adding one volume of 70% ethanol, the lysate was loaded onto the spin cartridge provided by the kit, washed, treated with DNAseI (PureLink DNase Set, Thermo Fisher Scientific, #12185010) and eluted in RNAse-free water.
cDNA synthesis was performed using SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific, #18080051), following the manufacturer's instructions.
Quantitative real-time PCR (qPCR) was performed with SYBR GreenER qPCR SuperMix Universal (Thermo Fisher Scientific, #11762500) using a CFX96 Real-Time PCR Detection System (Bio-Rad). Primers used for qPCR are listed in Supplementary Table S1. Relative quantification was performed using the ΔΔCt method.
Specific details of each data set are provided in the figure legends.
Luciferase assays
CHO or HeLa cells were seeded in 6-well plates and after 24 h transfected with 160 ng of 5X GAL4-UAS-firefly luciferase and 25 ng of pGL4.73[hRluc/SV40] Renilla luciferase (Promega, #E6911) vectors plus 350 ng of Gal4-λN in combination with 1000 ng of empty vector (EV) or BoxB-based constructs using Lipofectamine LTX Reagent with PLUS Reagent (Thermo Fisher Scientific, #15338100) or Lipofectamine 3000 (Thermo Fisher Scientific, #L3000150), respectively, following the manufacture instructions. To test the relevance of WDR5, WDR5 or non-silencing control siRNAs for loss of function, or 500 ng of pcDNA-STREP-HA-WDR5 or empty vector for gain of function, were cotransfected with some of the above plasmids as specified in individual Figures. As control of specificity, 160 ng of 5X GAL4-UAS-firefly luciferase and 25 ng of pGL4.73[hRluc/SV40] Renilla luciferase were also cotransfected with 350 ng of GAL4-VP16 in combination with WDR5 or non-silencing control siRNAs. For all transfection, the total DNA amount was maintained at 2000 ng using the corresponding empty vectors, when required. 48h post-transfection, cells were harvested, washed once in PBS, and firefly and Renilla luciferase activities were measured using the Dual-Reporter Luciferase Assay System (Promega, #E1910), according to the manufacturer instructions. Briefly, each well of the 6-well plate was lysed in 150 μl of Passive Lysis Buffer and incubated for 20 min at room temperature by gently shacking. Samples were then collected into 1.5 ml Eppendorf tubes and centrifugated for 10 min at 16 000g. The supernatant was divided in 3 independent wells of a 96-well plate opaque flat bottom and the values of each firefly and Renilla intensities were measured independently and subsequently displayed as an average value. The luminescence was acquired and quantified by Wallac 1420 multilabel Victor3 microplate reader (Perkin Elmer). Firefly luciferase data were normalized to Renilla luciferase activity.
Protein extraction and immunoblotting
Whole cell lysates were performed using either IP buffer (50 mM Tris–HCl pH 7,5; 150mM NaCl; 1% NP-40; 5 mM EDTA; 5 mM EGTA; protease inhibitors) or Laemmli Buffer 4×. Protein extracts were resolved on SDS-PAGE polyacrylamide gels and transferred to nitrocellulose blotting membrane (GE Healthcare Life Sciences). Membranes were incubated with the following primary antibodies: α-FLAG M2 (Sigma-Aldrich, #F1804), α-STREP-HRP (Sigma-Aldrich, #RABHRP3), α-Vinculin (Sigma-Aldrich, #V9131), α-BirA (Abcam, #ab232732), α-WDR5 (Abcam, #ab56919). Secondary antibodies conjugated to horseradish peroxidase were used at 1:10 000 dilutions (Jackson Immunoresearch, anti-mouse IgG HRP #715–035-150, anti-rabbit IgG HRP #711-035-152). Signal was visualized using the ECL Western blotting substrate (Thermo Fisher Scientific, Waltham, MA).
Csy4-BirA DBE-T frag.3 interactome
For each construct, 1 × 10 cm dish of Flp-In T-REx HEK293 BirA/FLAG/NLS/Csy4 cells treated with doxycycline 1 μg/ml for 28 h and biotin 50 μM for 24 h was used. The cell pellet was resuspended in 500 μl IP buffer (50 mM Tris–HCl pH 7.5; 1% NP-40; 5 mM EDTA; 5 mM EGTA; protease inhibitors) with 750 mM NaCl and incubated 15 min on ice to extract proteins. Afterwards, the NaCl concentration was reduced to 150 mM using IP-buffer without NaCl. The extract was passed through a 5 ml syringe 21G needle for 6× times to disrupt DNA and homogenize the sample. Next, samples were digested with 100 U of benzonase, while rocking on a wheel for 20 min at 37°C. The samples were then centrifuged at 160 000g for 20 min at 4°C and the concentration of the protein extract was measured using a Bradford assay. The extracts were brought to a final concentration of 1 μg/μl using IP buffer (50 mM Tris–HCl pH 7,5; 150 mM NaCl; 1% NP-40; 5 mM EDTA; 5 mM EGTA; protease inhibitors). Next, 5 mg of protein extracts were incubated with 40 μl STREP-T-actin beads (Sigma, #GE28-9355-99) in IP buffer while rocking for 2 h at 4°C. Afterwards, the beads were washed 3× in IP buffer and then 2× in IP buffer without NP-40 and protease inhibitors. Proteins were eluted from the beads with 50 μl 2.5 mM biotin in IP buffer without inhibitors and NP-40 for 1h at 4°C in a thermomixer at 1100 rpm.
To fully elute biotinylated proteins, the beads were further incubated with 50 μl of 2% SDS buffer in 50 mM Tris–HCl pH 7.4 for 1 h at 37°C in a thermomixer at 1100 rpm. Both eluates were then combined, boiled at 95°C for 5 min and processed according to the fASP protocol (28).
Mass-spectrometry analysis and protein identification
Proteins were reduced with Dithiothreitol (DTT), alkylated with Iodocetamide and digested with sequencing grade Trypsin (MERCK). Tryptic peptides were desalted using StageTip C18 (Thermo Fisher Scientific) and analyzed by nLC-MS/MS using a Q-Exactive mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) equipped with a nano-electrospray ion source (Proxeon Biosystems) and a nanoUPLC Easy nLC 1000 (Proxeon Biosystems). Peptide separations occurred on a homemade (75 μm i.d., 12 cm long) reverse phase silica capillary column, packed with 1.9-μm ReproSil-Pur 120 C18-AQ (Dr Maisch GmbH, Germany). A gradient of eluents A (distilled water with 0.1% v/v formic acid) and B (acetonitrile with 0.1% v/v formic acid) was used to achieve separation (300 nl/min flow rate), from 0% B to 45% B in 45 min for the first biological replicates; from 5% B to 50% B in 88 min for the second biological replicates. Full scan spectra were acquired with the lock-mass option, resolution set to 70 000 and mass range m/z from 380 to 1800 for the first biological replicates; from 300 to 2000 for the second biological replicates. The ten most intense doubly and triply charged ions were selected and fragmented. To quantify proteins, the raw data were loaded into the MaxQuant software version 1.6.1.0 to search the human_proteome 20220525 (101676 sequences; 41413969 residues). Searches were performed with the following settings: trypsin as proteolytic enzyme; five missed cleavages allowed; carbamidomethylation on cysteine as fixed modification; protein N-terminus-acetylation, methionine oxidation as variable modifications. Peptides and proteins were accepted with a FDR lower than 1%. Label-free protein quantification was based on the spectral counts considering only proteins identified with a minimum of two peptides of which at least one must be unique.
Specific interactors have been defined as those proteins reproducibly identified in all the biological replicates in association with DBE-T Frag.3 RNA and never or with >2-fold enrichment compared to the control RNA.
Recombinant protein purification
GST and GST-WDR5 proteins were expressed in Rosetta2 (DE3) pLys Escherichia coli (Novagen). Bacteria were grown in LB medium supplemented with antibiotics and induced with 1 mM IPTG (Biosciences) for 3 h at 37°C. Bacterial pellets were resuspended in Lysis Buffer (PBS; 1 mM PMSF; 5 mM 2-mercaptoethanol) and sonicated using a Bandelin sonoplus HD3100 (probe MS73) sonicator (10 cycles of 30 s on and 30 s off 80% amplitude). Lysates were subsequently incubated by gentle rotation for 15 min at 4°C after adding 1% Triton X-100 (Sigma) and centrifuged at 16 000g at 4°C for 20 min. Supernatants containing proteins were incubated 1 h at 4°C with Glutathione-Agarose beads (Sigma, # G4510) to retrieve GST-tagged proteins. Beads were washed with Lysis Buffer and purified proteins were eluted with elution solution (20 mM glutathione, 100 mM Tris–HCl pH 8.0, 120 mM NaCl). Proteins were dialyzed overnight at 4°C in Slide-A-Lyzer Dialysis Cassettes (Thermo Fisher Scientific, # 66380) in PBS. The purification steps and the integrity of the obtained proteins were analyzed by Coomassie Blue staining loading samples on 10% SDS-PAGE polyacrylamide gels.
In vitro RNA transcription
For the transcription of the different DBE-T fragments, each pGEM-T vector was linearized, and the inserted cDNA transcribed in vitro by using T7 RNA polymerase (Promega, #P2075). For the generation of the antisense RNA, the Sp6 polymerase was instead used, according to the manufacturer instruction (Promega, # P1085). Transcripts were treated with RNase-free DNase (Promega, #M6101) for 1 h at 37°C, purified with TRIzol (Thermo Fisher Scientific, # 15596026), and treated with TURBO DNase (Thermo Fisher Scientific, #AM2238). To preserve the proper RNA folding, each RNA was initially denatured for 2 min at 95°C, then put on ice for 5 min and finally recovered in RNA-folding buffer (10 mM Tris pH 8.0, 100 mM NaCl, 6 mM MgCl2) for 20 min at room temperature, as previously described in (29).
In vitro RNA-protein interaction
For the in vitro pulldown assays, 40 pmol of recombinant purified GST or GST-WDR5 proteins were immobilized on Glutathione-Agarose beads (Sigma) and incubated with 0.5 pmol of in vitro transcribed RNA in PD buffer (2 mM MgCl2, 0.2 mM ZnCl2, 1 mM DTT, 100 U/ml RNAseA inhibitor, 0.05% BSA, 0.2% NP-40, 1× PBS) for 1 h at room temperature. Beads were washed three times with PD buffer supplemented with 150 mM NaCl and then resuspended in TRIzol (Thermo Fisher Scientific, # 15596026) to extract the bound RNA. The extracted RNA was then processed for RT-qPCR as reported above.
RNA immunoprecipitation
The assay was carried out mainly as described in (19). Briefly, cells were seeded in 15 cm dishes (1 for each RNA IP), transfected with BoxB-LacZ or BoxB-DBE-T Frag.3 vectors and, after 48 h, UV cross-linked with 100000 μJ/cm2 twice (interval of 1′ between the two irradiations) on ice. Cells were lysed in 0.5% NP40, 0.5% Na Deoxycholate, 300 U/ml SUPERase•In RNase Inhibitor (Thermo Fisher Scientific, # AM2696), and protease inhibitors in PBS pH 7.9 and put on rotation for 25 min at 4°C. Samples were treated with 30 U of TURBO DNase (Thermo Fisher Scientific, #AM2238) and incubated 15 min at 37°C. After centrifuging 5 min at 1350g at 4°C, the supernatant was used in RNA IP. For each RNA IP 100 μl of Dynabeads Protein G (Thermo Fisher Scientific, #10003D), with 10 μg of anti-WDR5 antibody (Abcam, #ab56919) or IgG (Jackson lab, #015-000-003) as control, were used. Before performing the RNA IP, 10% of the supernatant was saved as input. The antibody-conjugated beads were added to the samples and put on rotation at 4°C overnight. Beads were washed three times (each for 5 min at 4°C) with PBS supplemented with 1% NP40, 0.5% Na Deoxycholate, additional 150 mM NaCl (final 300 mM), and 1:200 SUPERase•In RNase Inhibitor. Beads were resuspended in 100 μl of PBS + DNase buffer and 3 U of TURBO DNase was added. Samples were incubated 30 min on rotation at 37°C. Beads were washed three times (each for 5 min at 4°C) with PBS supplemented with 1% NP40, 0.5% Na Deoxycholate, 10 mM EDTA, additional 150 mM NaCl (total 300 mM), and 1:200 SUPERase•In RNase Inhibitor. RNA was eluted with 100 μl of 100 mM Tris–HCl (pH 7.5), 50 mM NaCl, 10 Mm EDTA, 100 μg Proteinase K, 0.5% SDS for 30 min at 55°C, with shaking. Eluate was centrifuged at 16 100g at RT and the supernatant was collected. Samples were purified with TRIzo LS Reagent (Thermo Fisher Scientific, #10296010) following the manufacturer's instructions. After RNA resuspension in 21.5 μl of RNase-free water, samples were treated with TURBO DNase, purified and 5 μl of DNA-free RNA were used for RT-qPCR as reported above.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed essentially as described in (19). Briefly, cells were harvested, washed once in PBS and immediately fixed for 10min at room temperature in 1% formaldehyde (Sigma, #47608) in PBS. Afterwards, formaldehyde was quenched with 125 mM glycine for 5 min and cells washed three times for 5min in PBS with gentle swirl. Cells were collected with a silicon scraper, transferred into 50 ml tubes and centrifuged at 1350g for 5 min at 4°C. Each pellet was lysed in 5 ml of LB1 solution (50 mM HEPES–KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP40, 0.25% Triton X-100) and incubated 10 min on ice. Next, samples were centrifuged at 1350g for 5 min at 4°C. The resulting pellet was washed in 5 ml of LB2 solution (10 mM Tris–HCl pH 8.0, 200 mM NaCl, 1 mM EDTA, and 0.5 mM EGTA) with gentle swirl 10 min at room temperature. Afterwards, samples were centrifuged at 1350g for 5 min at 4°C and the resulting pellet was lysed in 3 ml of LB3 (10 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM EDTA and 0.5 mM EGTA, 0.1% Na-Deoxycholate, 0.5% N-laurylsarcosine). LB1, LB2 and LB3 were supplemented with protease inhibitors (Complete EDTA-free Protease Inhibitor Cocktail Tablets; Roche). Lysates were sonicated with Bioruptor (Diagenode) for 10 min (medium intensity 30″ on 30″ off). An aliquot (5%) of the sonicated material was collected to determine the quality of the chromatin which must be in a range of 300–500 bp. The samples were quantified with Nanodrop spectrophotometer to determine the concentration of chromatin. Before ChIP, Triton X-100 was added to chromatin samples at a final concentration of 1% and a clarification step of 10 min at 16 360g at 4°C was performed. Supernatants were transferred into a new tube and precleared with 1/50 vol of Dynabeads protein G (Invitrogen, #10003D) rotating for 2 h at 4°C. Hundred micrograms of chromatin were used for each ChIP. For each ChIP, 100 μl of Dynabeads protein G were washed three times with 0.5% BSA in PBS and incubated with 5 μg of anti-WDR5 antibody (Abcam, #ab56919) or IgG (Jackson lab, #015-000-003) antibodies as control for 2 h on rotation at 4°C. Next, the beads-antibody complex was washed three times with 0.5% BSA in PBS and resuspended in 50 μl of 0.5% BSA in PBS. Chromatin and beads-antibody complexes were mixed and rocked overnight at 4°C. The day after, before starting the washes, 5% of the total ChIP volume was taken from the control IgG supernatant and subsequently used as input fraction. Next, six washes of 5 min each rocking at 4°C in RIPA buffer (50 mM HEPES–KOH pH 7.6, 500 mM LiCl, 1 mM EDTA, 1% NP-40, 0.7% Na-Deoxycholate) were performed. An additional wash of 5 min on rotation at 4°C in TE buffer (10 mM Tris–HCl pH 8.0, 1 mM EDTA) with 50 mM NaCl was performed. Next, samples were centrifuged for 3 min at 1000g at 4°C and the beads-antibody-chromatin complexes were eluted with elution buffer (TE buffer with 2% SDS) in a thermo mixer for 15 min at 65°C, shaking, and then centrifuged for 1 min at room temperature at 16 360g. The eluted material was transferred to a new tube and samples and input fractions were cross-link reverted overnight at 65°C. Finally, samples were purified with the QIAquick PCR purification kit (QIAGEN, #K310001), following the manufacturer instruction. DNA was eluted in 70 μl of TE buffer and 2 μl were used in qPCR analysis.
Apoptosis assay
Apoptotic levels were assessed by the IncuCyte S3 Imager system (Sartorious, USA). Cell death was been quantified by the caspase-3/7 reagent (Sartorious, USA #4440), which generates a green fluorescent signal upon activation of apoptotic pathways. Following manufacture's protocol, the IncuCyte Caspase-3/7 dye was diluted 1:1000 in complete differentiating media for a final assay concentration of 5 μM. The amount of green signal was quantified after treatment with either siRNA or OICR during 72 h with an acquisition of 36 pictures for each well of a 12-well plate every 3 h. Measurements and analyses were performed with the IncuCyte S3 software (Sartorius, USA) by averaging the technical replicates and normalizing to the fluorescence detected at T = 0.
Immunofluorescence
96 h after differentiation, mature myotubes derived from FSHD primary myoblast were fixed with 4% paraformaldehyde for 10 min at 4°C temperature and washed with PBS. Cells were incubated with blocking buffer containing 5% Goat serum and 0.1% Triton X-100 for 1 h. Then, cells were incubated with primary antibody anti-MHC (Developmental Studies Hybridoma Bank, anti-myosin heavy chain, MF-20) for 45 min at RT, washed 3 times with PBS and incubated with secondary antibody (Thermo Fisher Scientific, goat anti-mouse Alexa fluor 488, #A32723) and DAPI for 45 min. Fluorescent images were taken by a Carl Zeiss AxioImage M2 fluorescent microscope (Zeiss, Germany). The differentiation index was calculated as the frequency (percentage) of nuclei inside Mhc-positive cells in comparison with the total number of nuclei. The fusion index was calculated as the frequency (percentage) of nuclei inside myotubes (Mhc-positive syncytia containing at least three nuclei) in comparison with the total number of nuclei. The nuclei distribution was calculated as the frequency (percentage) of Mhc-positive cells containing the indicated number of nuclei. Three independent differentiation experiments were performed and 5 fields per well were analyzed for each sample/experiment.
OICR-9429 treatment
FSHD and healthy donor muscle cells were induced to differentiate (see above). 60 h after induction of differentiation, cells were treated for 36 h with 20 μM of OICR-9429 (Sigma-Aldrich, #SML1209) or DMSO as control for a total 96 h of differentiation.
dsRNA-IP
Human muscle cells treated with WDR5 and non-targeting siRNAs were washed in Phosphate-Buffered Saline (PBS), trypsinized and counted prior to lysis. Approximately 1.2 × 106 cells were used for each IP as described in (30). Specifically, 1 ml total volume was sonicated using a Diagenode Bioruptor on light setting (5 min total, 30 s on/off at 4°C) in lysis buffer (15 mM Tris pH 7.5, 0.1 M NaCl, 5 mm MgCl2, 0.5% Triton X-100, 1 mm dithiothreitol) plus 40 U/ml RNase inhibitor (Ambion, #AM2682). 10% of the supernatant was saved as input. Each lysate was precleared using 40 μl of protein G Dynabeads (Invitrogen, #10003D) for 1 h prior to an overnight incubation at 4°C with either K1 (Jena Bioscience, #RNT-SCI-10020200) or an IgG (Jackson lab, #015-000-003) control antibodies. Then 40 μl of protein G Dynabeads (Invitrogen, #10003D) were added for 1 h to bind the antibody, and beads were subsequently washed four times with 1 ml of cold lysis buffer. After the final wash, 1 ml of TRIzol (Thermo Fisher Scientific, # 15596026) to extract the bound RNA. The extracted RNA was then processed for RT-qPCR as reported above.
Prime-seq
Prime-seq was carried out essentially as previously described in (31). A step-by-step protocol, including all materials, is available on protocols.io (https://doi.org/10.17504/protocols.io.s9veh66).
Total RNA was isolated from six biological replicates for each sample by employing the PureLink RNA Mini Kit (Thermo Fisher Scientific, #12183020) including digestion of remaining genomic DNA according to producer′s guidelines. RNA was quantified using the Qubit RNA High Sensitivity Assay kit (Thermo Fisher Scientific, #Q32855).
20 ng of RNA from each sample was incubated with the reverse transcription mix, consisting of 30 units Maxima H- enzyme (Thermo Fisher Scientific, #EP0753), 1× Maxima H- Buffer (Thermo Fisher Scientific), 1 mM each dNTPs (Thermo Fisher Scientific), 1 μM template-switching oligo and 1 μM barcoded oligo (dT) primers. The reaction was incubated at 42°C for 90 min.
Following cDNA synthesis, the samples from the same donor (FSHD or healthy donor) were pooled, cleaned, and concentrated with AMPure XP beads (Beckman Coulter, #A63881) at a 1:1 ratio and eluted in 17 μl of nuclease-free water. Residual primers were digested using Exonuclease I (Thermo Fisher Scientific, #EN0581) at 37°C for 20 min followed by a heat inactivation step at 80°C for 10 min. The samples were cleaned using AMPure XP beads at a 1:1 ratio and eluted in 20 μl of nuclease-free water.
Second-strand synthesis and pre-amplification were performed in a 50 μl reaction, consisting of 1× KAPA HiFi Ready Mix (Roche, #7958935001) and 0.6 μM SingV6 primer, with the following PCR setup: initial denaturation at 98°C for 3 min, denaturation at 98°C for 15 s, annealing at 65°C for 30 s, elongation at 68°C for 4 min, and a final elongation at 72°C for 10 min. Denaturation, annealing, and elongation were repeated for 11 cycles.
The resulting DNA was cleaned using AMPure XP beads at a ratio of 1:0.8 of DNA to beads and eluted with 10 μl of nuclease-free water. The DNA quantity was assessed using Qubit DNA High Sensitivity Assay kit (Thermo Fisher Scientific, #Q32854) and the quality was assessed using an Agilent TapeStation with a High-Sensitivity DNA 5000 screen tape (Agilent, #5067-5592).
Libraries were prepared with the NEBNext Ultra II FS Library Preparation Kit (NEB, #E6177S) according to the manufacturer instructions with the exception of adapter sequence and reaction volumes. Fragmentation was performed on 2.5 μl of cDNA (generally 800 ng) using Enzyme Mix and Reaction buffer in a 6 μl reaction. A custom prime-seq adapter (1.5 μM) was ligated using the Ligation Master Mix and Ligation Enhancer in a reaction volume of 12.7 μl. The samples were then double-size selected using AMPure XP beads with a high cutoff of 0.5 and a low cutoff of 0.7. The samples were then amplified using Q5 Master Mix (NEB, #M0544L), 1 μl i7 Index primer and 1 μl i5 Index primer using the following PCR conditions: 98°C for 30 s; 11–12 cycles of 98°C for 10 s, 65°C for 1 min 15 s, 65°C for 5 min; and 65°C for 4 min. Double-size selection was performed once more as before. The quantity and quality of the libraries were also assessed as before.
Paired-end sequencing was performed on an Illumina NextSeq 2000 instrument.
Prime-seq data analysis
The zUMIs pipeline (described in (32), version 2.9.7), was used to filter the raw data, with a Phred quality score threshold of 20 for 2 barcode bases and 3 UMI bases. The filtered data was mapped to the human genome (GRCh38 Ensembl v105) using STAR (version 2.7.3a) (33) and the reads counted using RSubread (version 1.32.4) (34) by applying a downsampling to the sample with the lowest number of reads.
The count matrix generated by zUMIs (UMI counts in exons) was loaded into R (version 4.1.0, R Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2016. Available from: https://www.r-project.org/) and DESeq2 (version 1.32.0) (35) was used for differential gene expression analysis (adjusted p-value < 0.05). Only genes with UMI counts >1 were considered.
Gene set enrichment analysis (GSEA) of DUX4 target genes (36) was performed with fgsea R-package (version 1.18.0) (37).
The msigdbr R-package (version 7.5.1, msigdbr: MSigDB Gene Sets for Multiple Organisms in a Tidy Data Format. R package version 7.5.1, https://igordot.github.io/msigdbr/) with hallmark gene sets (http://www.gsea-msigdb.org/gsea/msigdb/collections.jsp#H) was used for the analysis of regulated pathways.
For data visualization was used ggplot2 (version 3.3.5, Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer; 2010).
Differentially regulated genes common to FSHD and healthy donor muscle cells treated with OICR-9429 were assigned to GO groups (Annotation data sets: PANTHER GO-Slim Biological Process and PANTHER Protein Class) using the PANTHER Overrepresentation Test (Released 20220712) (38) with default parameters and with all human genes as background.
Venn diagrams were generated with BioVenn (39). Statistics on the Venn diagrams was performed with a program developed by Jim Lund (University of Kentucky, Department of Biology) http://nemates.org/MA/progs/overlap_stats.html.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 9 (GraphPad Software, San Diego, USA). Statistical significance between groups was calculated by Student's t-test on at least three independent experiments. Differences among groups were assessed by Tukey-adjusted analysis of variance (ANOVA). P-value: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Data are expressed as mean ± standard deviation (SD). Details of each dataset are provided in the corresponding figure legend.
RESULTS
A reporter system to study DBE-T activity in mammalian cells
DBE-T is a lncRNA transcribed from a region immediately proximal to the D4Z4 repeat array and covers the first two centromeric repeats (19) (Supplementary Figure S1A). Dissecting the activity of the endogenous DBE-T is complicated since it is chromatin-associated, it works in cis only at the FSHD locus and, like DUX4, it is expressed only by a minority of nuclei (19,40). Moreover, ectopic DBE-T overexpression is unable to affect transcription (19), possibly because it is not correctly localized. To test whether DBE-T recruitment to a target gene is sufficient for its activation, we took advantage of the Gal4-λN-BoxB targeting system (26). The system is composed by: I. a firefly Luciferase reporter gene under the control of five tandem upstream binding sites specifically recognized by the yeast transcription factor GAL4 (5X GAL4-UAS Luciferase); II. the GAL4 DNA binding domain fused to the RNA-binding domain of the lambda bacteriophage antiterminator protein N (GAL4-λN); BoxB (containing five repeats of the λN-specific binding site) fused to either DBE-T or LacZ, as negative control. This system allows an RNA of interest to be recruited upstream of the Luciferase gene by GAL4-λN (Figure 1A).
Cells were co-transfected with the Luciferase and the GAL4-λN plasmids in combination with the BoxB-DBE-T, BoxB-LacZ or empty vector plasmids. In these setting, BoxB-DBE-T significantly promoted the expression of the Luciferase reporter as compared to BoxB-LacZ or the empty vector controls (Figure 1B). Several additional controls supported the specificity of findings. For example, DBE-T was unable to activate the reporter when not fused to BoxB (Supplementary Figure S1B). In addition, DBE-T silencing reduced BoxB-DBE-T activity (Supplementary Figure S1C). All in all, these results demonstrated that DBE-T works as an RNA molecule to activate gene expression.
Identification of a DBE-T functional domain
To determine if there is a DBE-T region mainly responsible for gene activation, we divided it in seven fragments of similar size that were all fused to BoxB (Figure 1C). Interestingly, only BoxB-Frag.3 was able to significantly activate the reporter gene expression (Figure 1D). Moreover, a BoxB-DBE-T mutant lacking the Frag.3 region significantly reduced the reporter activation (Figure 1E). Conversely, deletion of another DBE-T region (Frag.5) did not altered DBE-T-mediated gene activation (Figure 1E). In addition, the antisense of Frag.3 fused to BoxB failed to activate expression of the reporter (Figure 1F). Similar results were obtained by using the Gal4-λN-BoxB targeting system in a different cell line (Supplementary Figure S1D-E).
Collectively, our results indicate that the Frag.3 region of DBE-T is mainly responsible for gene activation.
Identification of DBE-T frag.3 interactors
RNA–protein interactions are involved in various cellular processes. We hence hypothesized that one or more protein factors associate to DBE-T Frag.3 to mediate gene activation. To isolate them, we established a new RNA-targeting system coupled with proximity-dependent biotinylation to identify RNA–protein interactions in intact cells. The system relies on the activity of the two previously described bacterial enzymes, Csy4* and BirA*. Csy4* is a mutant of the CsyA endonuclease part of the CRISPR (clustered regularly interspaced short palindromic repeats) system and stably binds to transcripts carrying a 16-nt Csy4 hairpin RNA-tag with very high affinity and specificity (25,41). BirA* is a mutant of the BirA protein biotin ligase that biotinylates proteins in a proximity-dependent manner (42). We reasoned that a fusion protein BirA*-Csy4* would be able to biotinylate proteins associated to a transcript of interest fused to the Csy4 RNA-tag. Biotinylated proteins could subsequently be isolated and identified by mass spectrometry (Figure 2A). To this aim, we generated HEK293-Flp-In cells that expressed, upon doxycycline administration, a BirA*-Csy4* version carrying the SV40 nuclear localization sequence (NLS) at its N-terminus. As shown in Supplementary Figure S2, the cells correctly expressed the enzymes only in the presence of doxycycline and biotinylated proteins once biotin was provided into the medium. We then expressed in these cells Csy4 hairpin tagged DBE-T Frag.3 (Csy4-Frag.3), the antisense of Frag.3 (Csy4-Frag.3 As) or just the Csy4 tag. Doxycycline and biotin were added directly into the medium 24 h after transfection and cells were harvested 24h after doxycycline/biotin administration. We then extracted proteins, isolated biotinylated proteins by Streptavidin beads and specifically eluted them using biotin competition and SDS elution (Figure 2A). The material was then In-solution digested and tryptic peptides analyzed by mass spectrometry in biological replicates. The Csy4-Frag.3 interactome was then filtered using the proteins present in the Csy4-empty vector or Csy4-Frag.3 As purifications. We retained only proteins that were >2-fold enriched in the Csy4-Frag.3 compared to both negative control interactomes (Figure 2B and Supplementary Table S2). Among them is WDR5, which is mostly known as core subunit of MLL/SET1 histone methyltransferase complexes. WDR5 is required for histone H3 lysine 4 trimethylation (43), which is enriched at the FSHD locus when DUX4 expression is activated (19). Moreover, WDR5 binding to specific lncRNAs is essential to maintain active chromatin (44). Hence, despite other proteins specifically associated to DBE-T Frag.3 could contribute to DBE-T activity, we decided to focus on WDR5.
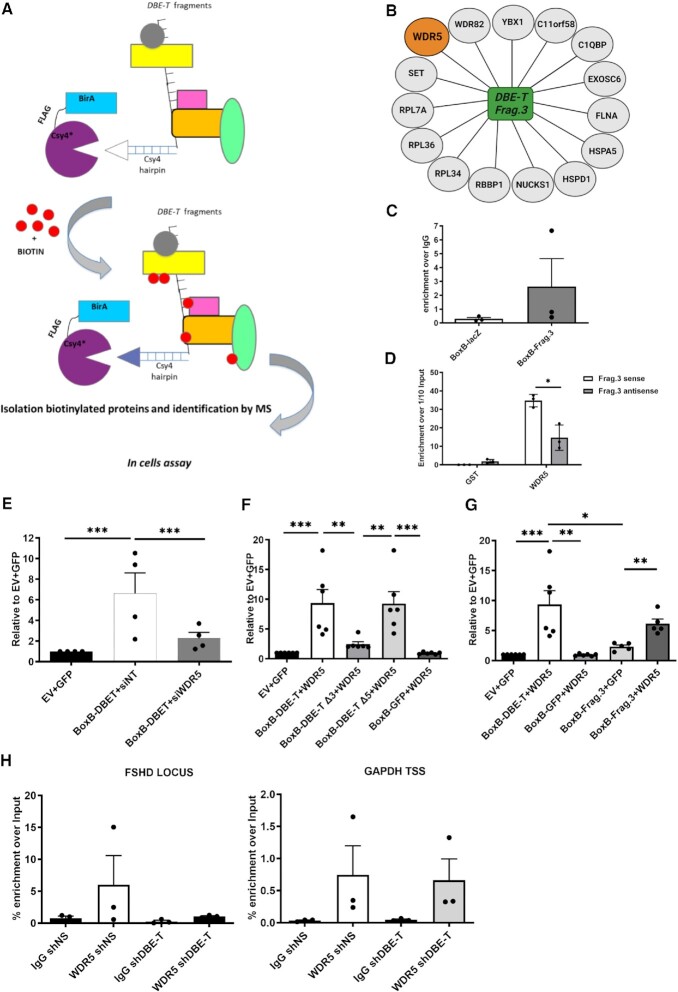
Figure 2.
Identification of DBE-T Frag.3 interactors. (A) Schematic representation (not in scale) of the RNA-targeting systems coupled with proximity-dependent biotinylation. The fusion protein BirA*-Csy4* biotinylates proteins associated to the RNA fused to the Csy4 RNA-tag. Then, biotinylated proteins are isolated and identified by mass spectrometry. (B) Schematic representation of the Csy4-DBE-T Frag.3 specifically associated proteins. (C) RNA immunoprecipitation following UV crosslinking (UV-RIP) of CHO cells transfected with BoxB-LacZ or BoxB-DBE-T using anti-WDR5 antibodies or IgG (control). The extracted RNA was processed for RT-qPCR showing the enrichment of DBE-T Frag.3 over the control LacZ RNA. N = 3. (D) In vitro pull-down experiments using purified, recombinant GST-WDR5 and in vitro transcribed DBE-T Frag.3 showing the direct interaction between GST-WDR5 and DBE-T Frag.3 and the significant preference for interacting with DBE-T Frag.3 compared to its antisense control. N = 4. Student's t-test. *P< 0.05. (E) WDR5 depletion caused a significant decrease in reporter gene activation by DBE-T. N = 4. Student's t-test. ***P< 0.001. (F) Gal4-λN-BoxB reporter assay showing WDR5 overexpression significantly boosted reporter gene activation by DBE-T and a significant reduction using a DBE-T version deleted of Frag.3, while a DBE-T mutant lacking Frag.5 is equally stimulated by WDR5 overexpression. N = 6. Student's t-test. **P< 0.01, ***P< 0.0001. (G) Gal4-λN-BoxB reporter assay showing WDR5 can specifically stimulate gene activation by DBE-T Frag.3. N = 5. Student's t-test. *P< 0.05, **P< 0.01, ***P< 0.001. (H) ChIP-qPCR showing WDR5 enrichment at the FSHD locus that is reduced upon DBE-T knockdown while WDR5 enrichment at the control locus GAPDH was unaffected by DBE-T knockdown. N = 3.
To validate WDR5 interaction with DBE-T Frag.3, we performed RNA immunoprecipitation following UV crosslinking (UV-RIP) with anti-WDR5 antibodies or IgG, as control. DBE-T Frag.3 was enriched in the WDR5 UV-RIP, while the control LacZ RNA was not (Figure 2C). Since UV irradiation identifies protein–nucleic acid interactions only when in close proximity (45), a direct WDR5-DBE-T Frag.3 binding in cells was suggested. To confirm this, we performed in vitro pull-down experiments using purified, recombinant GST-WDR5 and in vitro transcribed DBE-T Frag.3. GST-WDR5 was able to directly interact with DBE-T Frag.3 while no enrichment was obtained using GST alone. Moreover, GST-WDR5 displayed significant preference for interacting with DBE-T Frag.3 compared to its antisense control (Figure 2D). Hence, WDR5 binds directly and specifically to DBE-T Frag.3.
WDR5 is required for gene activation by DBE-T
To investigate the functional relevance of the WDR5/DBE-T interaction, we initially took advantage of the Gal4-λN-BoxB reporter assay. WDR5 depletion caused a significant decrease in reporter gene activation by DBE-T (Figure 2E). Importantly, using a modified system in which expression of the reporter is driven by the GAL4 DNA binding domain fused to the transcriptional activation domain of Herpes simplex virus Type 1 protein VP16 (46), the reporter gene activation was unaffected by WDR5 depletion (Supplementary Figure S3), in line with the fact that WDR5 is not required in general for gene activation.
WDR5 overexpression significantly boosted reporter gene activation by DBE-T (Figure 2F). This requires Frag.3 since the effect of WDR5 overexpression was significantly reduced using a DBE-T version deleted of Frag.3 (Figure 2F), while activity of a DBE-T mutant lacking Frag.5 was equally stimulated by WDR5 overexpression (Figure 2F). Accordingly, WDR5 was able to specifically stimulate gene activation by DBE-T Frag.3 alone (Figure 2G). Hence, WDR5 binding to Frag.3 is required for gene activation by DBE-T.
WDR5 is recruited to the FSHD locus by DBE-T
Having found that WDR5 binds directly to DBE-T and is required for gene activation by DBE-T, we wondered if WDR5 is associated to the FSHD locus and, if so, if DBE-T is required for this association. To test this, we performed chromatin immunoprecipitation followed by quantitative real-time PCR (ChIP-qPCR) using chromatin from terminally differentiated human FSHD muscle cells in combination with antibodies against WDR5 or IgG, as control. We used chromatin from FSHD myotubes expressing non-silencing (shNS) or DBE-T silencing (shDBE-T) shRNAs. Interestingly, we found that WDR5 is enriched at the FSHD locus and its enrichment is reduced upon DBE-T knockdown (Figure 2H). We previously showed that DBE-T is a lncRNA acting in cis only at the FSHD locus (19). Accordingly, WDR5 enrichment at the control locus GAPDH was unaffected by DBE-T knockdown (Figure 2H). Hence, WDR5 is specifically recruited to the FSHD locus in a DBE-T dependent manner.
WDR5 is required for DUX4 expression in FSHD
In FSHD muscle cells, both DUX4 and DBE-T expression are induced during myogenic differentiation (47–49), which we found shows a significant positive correlation (Supplementary Figure S4A–C). We analyzed the expression of WDR5 during myogenic differentiation of primary muscle cells derived from FSHD patients. Intriguingly, we found that WDR5 is significantly increased during differentiation (Supplementary Figure S4D). In line with the possibility that WDR5 might contribute to the myogenic activation of DUX4 we found a positive correlation between WDR5 expression and DUX4 (Supplementary Figure S4E). To test the relationship between WDR5 and DUX4, we transfected primary muscle cells isolated from independent FSHD patients with control (siNT) or WDR5 targeting (siWDR5) siRNAs (Supplementary Figure S5A, B). Depletion of WDR5 significantly reduced the expression level of DUX4 compared to the control (Figure 3A and Supplementary Figure S6A).
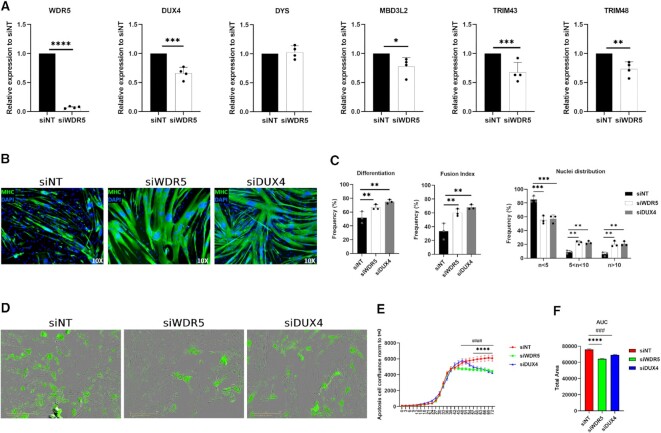
Figure 3.
Effects of WDR5 silencing with siRNA. (A) RT-qPCR for the indicated genes performed on RNA extracted from primary FSHD muscle cells treated with control (siNT) or WDR5 (siWDR5) siRNAs. N = 4. Student's t-test. *P< 0.05, **P< 0.01, ***P< 0.001, ****P< 0.0001. (B) Representative images of Myosin Heavy-chain (MHC, green) and Nuclei (DAPI, blue) staining performed on primary FSHD muscle cells treated with control (siNT), WDR5 (siWDR5) or DUX4 (siDUX4) siRNAs. (C) Quantification of differentiation index, fusion index and nuclei distribution in primary FSHD muscle cells treated with control (siNT), WDR5 (siWDR5) or DUX4 (siDUX4) siRNAs. No statistical difference between siWDR5 and siDUX4 treated cells was observed. N = 3. One-way ANOVA. **P< 0.01, ***P< 0.001. (D) Representative images obtained from the IncuCyte S3 Imager system monitoring live caspase 3/7 apoptosis. Images were acquired at 10× magnification in bright field. The apoptotic cells (green) were acquired with 300 ms exposure. (E) Analysis of the live-cell, real-time, caspase 3/7 apoptosis assays on primary FSHD muscle cells treated with siWDR5, siDUX4 or siNT (control). N = 3. One-way ANOVA. ####P< 0.0001, ****P< 0.0001. (F) Area under the curve (AUC) quantification. N = 3. Student's t-test. ###P< 0.001, ****P< 0.0001.
Since expression of DUX4 is restricted to differentiated FSHD muscle cells, treatments that reduce the differentiation capability of the cells might non-specifically decrease DUX4 expression. Importantly, WDR5 knockdown did not cause any significant reduction in the expression of Dystrophin (Figure 3A and Supplementary Figure S6A), which is mainly expressed in differentiated myotubes (50,51) like DUX4, suggesting that WDR5 silencing does not inhibit myogenesis (see also below).
Given that the ability of DUX4 to activate gene expression is strictly required to execute its toxicity (52–58), we analyzed expression of known DUX4 targets. Intriguingly, we found a significant decrease in the expression of DUX4 targets upon WDR5 silencing compared to control knockdown (Figure 3A and Supplementary Figure S6A).
WDR5 silencing rescues FSHD muscle cell differentiation
DUX4 expression has been shown to interfere with muscle differentiation (59) and muscle cells from FSHD patients display impaired myogenesis (60–62). We treated primary muscle cells isolated from independent FSHD patients with control or WDR5 siRNAs and measured their ability to differentiate into myotubes. As shown in Figure 3B, C and Supplementary Figure S6B, C, we found that WDR5 knockdown significantly rescues the myogenic and fusion indexes of FSHD muscle cells allowing to produce myotubes with a significantly increased number of myonuclei with respect to control knockdown cells (fibers with more than 10 nuclei from 6.02% ±2.89SD to 20.39% ±4.24SD). As positive control, we performed DUX4 knockdown. Remarkably, we found no statistical difference in the rescue of myogenesis caused by direct DUX4 knockdown compared to WDR5 silencing (Figure 3C), further supporting the relevance of WDR5 in FSHD.
WDR5 silencing rescues FSHD muscle cell viability
DUX4 expression in multiple cell lines as well as in skeletal muscle in vivo leads to apoptotic cell death (47,52,53,55,59,63). Increased apoptosis and its dependence on DUX4 has been documented in FSHD muscle cells and tissues (47,64–66).
In primary muscle cells of FSHD patients, DUX4 is expressed by a minority of FSHD myonuclei (40,67). Hence, only a fraction of FSHD muscle cells undergoes DUX4-induced cell death (47) making it difficult to monitor the efficacy of possible therapeutic treatments. To address this issue, we performed live-cell, real-time, caspase 3/7 apoptosis assays in thousands of cells from each culture in an automated and unbiased manner. This approach allows to correlate apoptotic signals with high-definition phase contrast images to provide additional biological insight and morphological validation of apoptotic cell death (e.g. cell shrinkage, membrane blebbing, nuclear condensation). To test the ability of WDR5 silencing to protect from endogenous DUX4-induced cell death, we transfected primary muscle cells isolated from independent FSHD patients with control or WDR5 targeting siRNAs and monitored cell death over time. As shown in Figure 3D–F and Supplementary Figure S6D–F, WDR5 knockdown leads to a significant decrease in FSHD muscle cell death with respect to control knockdown cells. As positive control, we performed DUX4 knockdown (Supplementary Figure S7). Notably, we found no statistical difference in the rescue of muscle cells viability caused by direct DUX4 knockdown compared to WDR5 silencing.
Importantly, WDR5 knockdown did not affected myogenic differentiation or viability of primary muscle cells derived from independent healthy donors (Supplementary Figure S8A–F).
To date, the only direct DUX4 transcriptional target required for DUX4-induced cell death is human satellite II (HSATII) through the production of toxic dsRNA (30). For this reason, we analyzed the expression levels of HSATII upon WDR5 targeting. Intriguingly, WDR5 knockdown in primary FSHD muscle cells caused a significant reduction of HSATII levels compared to the respective controls (Supplementary Figure S9A). Moreover, the overall amount of HSATII dsRNA was significantly reduced upon WDR5 silencing (Supplementary Figure S9B).
Collectively, our data indicate that WDR5 silencing rescues viability and myogenic differentiation of FSHD muscle cells without any obvious effect on healthy muscle cells.
WDR5 pharmacological targeting inhibits DUX4 expression and reduces FSHD muscle cell pathologic signs
WDR5 is a highly conserved protein involved in a variety of processes ranging from gene regulation to the control of cell division (68). Hence, targeting WDR5 expression would not be a valuable therapeutic strategy for FSHD. Intriguingly, WDR5 displays several binding pockets that can be selectively targeted with small molecule inhibitors. Most available drugs target the WIN site of WDR5, which mediates the interaction with chromatin and the MLL/SET enzymes. Importantly, WIN site inhibitors affect only a specific subset of WDR5 activities making them more attractive for clinical applications (69).
We decided to test OICR-9429, a commercially available small-molecule WIN site antagonist of the interaction between WDR5 and the MLL/SET1 methyltransferase complexes (70). Pharmacological inhibition of WDR5 by treating primary FSHD muscle cells with OICR-9429 significantly reduced WDR5 association with the FSHD locus (Figure 4A), leading to a significantly reduced expression of DUX4 and its targets without reducing the levels of the control gene Dystrophin (Figure 4B). Moreover, OICR-9429 treatment caused a significant amelioration of differentiation and fusion index coupled with a significant increase in multinucleated myofibers as compared to DMSO treated controls (Figure 4C, D). In addition, WDR5 inhibition led to a significant decrease in FSHD muscle cell death with respect to control treated cells (Figure 4E–G).
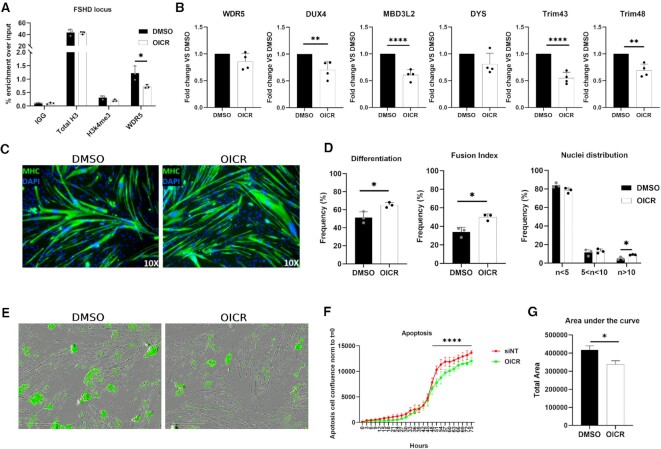
Figure 4.
Effects of WDR5 pharmacological inhibition with OICR-9429. (A) ChIP-qPCR showing WDR5 enrichment at the FSHD locus that is significantly reduced upon WDR5 pharmacological inhibition with OICR9429. (B) RT-qPCR for the indicated genes performed on RNA extracted from primary FSHD muscle cells treated with OICR-9429 or DMSO (as control). N = 4. Student's t-test, **P< 0.01, ****P< 0.0001. (C) Representative images of Myosin Heavy-chain (MHC, green) and Nuclei (DAPI, blue) staining performed on primary FSHD muscle cells treated with DMSO (control) or OICR-9429. (D) Quantification of differentiation index, fusion index and nuclei distribution in primary FSHD muscle cells treated with DMSO (control) or OICR-9429. N = 3. Student's t-test. *P< 0.05. (E) Representative images obtained from the IncuCyte S3 Imager system monitoring live caspase 3/7 apoptosis. Images were acquired at 10× magnification in brighfield. The apoptotic cells (green) were acquired with 300ms exposure. (F) Analysis of the live-cell, real-time, caspase 3/7 apoptosis on primary FSHD muscle cells treated with OICR-9429 or DMSO, as control. The analysis was performed with the IncuCyte S3 software. N = 4. One-way ANOVA. ****P< 0.0001. (G) Area under the curve (AUC) quantification. N = 4. Student's t-test. *P< 0.05.
Notably, WDR5 pharmacological inhibition did not affected myogenic differentiation or cell viability of primary muscle cells derived from independent healthy donors (Supplementary Figure S10A–F).
Pharmacological WDR5 targeting restores DUX4-mediated gene alterations
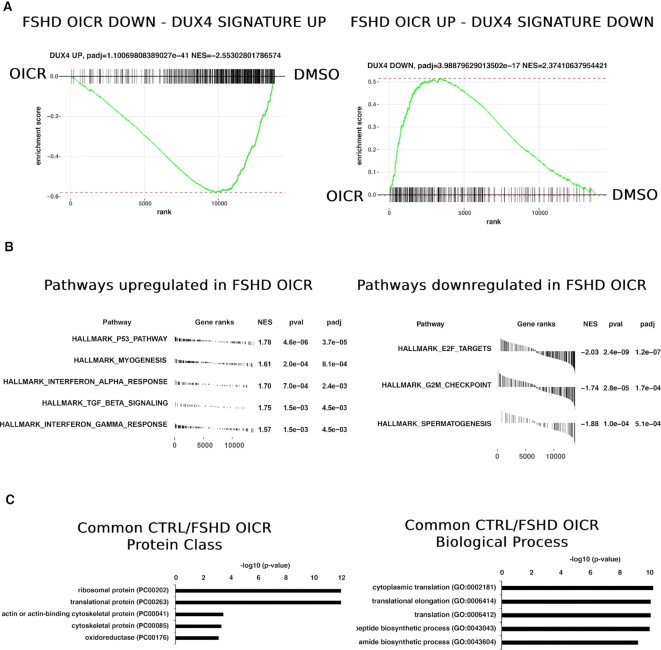
To analyze the genome-wide effect of WDR5 pharmacological inhibition, we performed transcriptomic analyses through RNA-seq to compare primary FSHD or healthy donor muscle cells treated with OICR-9429 or DMSO, as control. (Supplementary Table S3). Importantly and selectively in FSHD muscle cells, WDR5 inhibition caused an evident impact on the DUX4-dependent gene expression (Figure 5A and Supplementary Figure S11A), which is the major molecular signature in FSHD skeletal muscle (55). Gene set enrichment analysis (GSEA) based on DUX4 regulated genes (57) showed a significant overall increase in FSHD-OICR muscle cells of the genes that have been reported to be downregulated upon DUX4 expression and, vice versa, a global and significant downregulation of the genes that have been reported upregulated when DUX4 is expressed (Figure 5A). Moreover, selectively in FSHD muscle cells the myogenesis gene set was significantly upregulated while cell cycle gene sets were significantly downregulated (Figure 5B), in line with the rescue of myogenic differentiation observed by OICR-9429 treatment (Figure 4C-D). Instead, genes commonly altered in primary FSHD and healthy donor muscle cells treated with OICR-9429 were significantly enriched for genes encoding for ribosomal proteins or proteins involved in translation (Figure 5C, Supplementary Figure S11B and Supplementary Table S4), in line with previous data showing that a conserved effect of WDR5 inhibition is the alteration of genes connected to protein synthesis (71).
Figure 5.
Pharmacological WDR5 targeting restores DUX4-mediated gene alterations. (A) GSEA (Gene set enrichment analysis) performed on the DUX4-associated gene signature. Only in primary FSHD muscle cells treated with OICR-94289, there is a significant overall increase of the genes that have been reported to be downregulated upon DUX4 expression and, vice versa, a global and significant downregulation of the genes that have been reported upregulated when DUX4 is expressed. NES: normalized enrichment score. (B) Hallmark gene sets analysis showing the pathways that are significantly up- or down-regulated selectively in primary FSHD muscle cells after treatment with OICR-9429. (C) Gene Ontology analysis performed on genes commonly altered in FSHD and healthy donor primary muscle cells treated with OICR-9429. N = 6. Only the top 5 GO terms (ranked by P-value) are shown. Significance shown in the figures.
Collectively, our expression profiling indicates that WDR5 pharmacological inhibition causes a significant effect on the expression of DUX4-regulated genes and offers molecular support to our functional results.
DISCUSSION
FSHD differs from most Mendelian disorders for the peculiar genetic defects underlying the disease and the strong role of epigenetic components. The disease is linked to copy number reduction and/or epigenetic alterations of the D4Z4 macrosatellite on chromosome 4q35 and associated with loss of DUX4 epigenetic silencing leading to its reactivation, which is toxic to muscle cells. While preventing DUX4 aberrant expression is a plausible therapeutic option for FSHD patients, we know relatively little of the factors required for DUX4 expression in FSHD muscle cells.
Collectively, our data indicate that WDR5 plays a key role in the aberrant DUX4 re-activation and its pathological effects in FSHD muscular dystrophy. We found that WDR5 binds directly and specifically to the activating lncRNA DBE-T, which is responsible for WDR5 recruitment to the FSHD locus. Genetic or pharmacological WDR5 inhibition significantly reduce DUX4 expression, ameliorating several pathological signs displayed by muscle cells of FSHD patients. Importantly, similar treatments do not cause any appreciable phenotypic change in muscle cells from healthy donors.
To have a global view of the effects caused by WDR5 inhibition, we performed RNA-seq on FSHD or healthy donor muscle cells treated with the WDR5 inhibitor OICR-9429. As expected, we found that a relatively small group of genes encoding for ribosomal proteins is commonly affected in FSHD and control muscle cells in line with previous results showing downregulation of specific protein synthesis genes in all cell lines treated with WDR5 inhibitors (71). Importantly, selectively in FSHD muscle cells, WDR5 inhibition has a profound effect on the expression of DUX4-regulated genes which constitute the major molecular signature in FSHD skeletal muscle (55,57) and are used as biomarkers of disease progression and response to therapy in current clinical trials (72,73).
Our expression profiling provides also molecular support to our functional results. Previous studies indicate that DUX4 inhibits muscle differentiation by downregulating the myogenic transcription factor MyoD (59,74). Intriguingly, the significant upregulation of MyoD expression in primary FSHD muscle cells by WDR5 pharmacological inhibition (Supplementary Table 3) likely contributes to the amelioration of myogenic differentiation that we documented upon WDR5 targeting. Concerning apoptosis, aberrant activation of bi-directional HSATII transcription leading to dsRNA formation was shown to significantly contribute to DUX4 toxicity (30). We found that WDR5 inhibition reduces HSATII dsRNA, which is likely part of the mechanism through which WDR5 targeting significantly reduces FSHD muscle cells apoptotic levels.
The lncRNA DBE-T is expressed by few nuclei, mainly in terminally differentiated muscle cells of FSHD patients like DUX4, and function in cis just at the FSHD locus chromatin (this study and (19). These features make it very complicated to dissect the activity of the endogenous DBE-T. To circumvent these limitations and focus just on the activity of DBE-T, we adapted the Gal4-λN-BoxB targeting system (26). Our results strongly indicates that the DBE-T RNA molecule, rather than the act of its transcription, activates DUX4 expression at the FSHD locus.
To identify proteins associated to DBE-T in living cells, we developed a new CRISPR-based RNA proximity proteomic approach coupling the BirA protein biotin ligase and the Csy4 nuclease (25,41,42). This RNA-centric method could potentially be extended to other RNAs to identify proteins co-factors in a native cellular context.
WDR5 is a multitasking protein involved in the assembly of several histone-modifying and gene regulation complexes. While it is primarily known for its regulatory role in the MLL/SET complexes that deposit histone H3 lysine 4 (H3K4) methylation and the non-specific lethal (NSL) complex responsible for histone H4 lysine 16 acetylation, WDR5 also interacts and regulates the activity of the MYC and the retinoic acid receptor transcription factors, the kinesin motor protein Kif2A, the protein kinase PDPK1, and several others (68,75). Understanding which WDR5 containing complex or histone modification plays a major role in the activation of DUX4 gene expression in FSHD is highly relevant. Nevertheless, the moonlighting roles of WDR5 make this goal for the future very challenging. Related to this, while for the present study we focused on WDR5, additional candidates identified by our BirA*-Csy4* targeting system could collaborate with WDR5 to the regulation of DUX4 expression. Future work is required to establish if they are part of the same complex recruited by DBE-T to activate DUX4 in FSHD patients.
Given its centrality in FSHD, possible therapeutic approaches could target DUX4 expression, activity or downstream pathways. Inhibition of transcription factors activity is notoriously difficult to achieve given their lack of small molecule inhibitor-friendly pockets. While targeting DUX4 downstream pathways would be a viable option, it is currently unclear which of them is responsible for the disease and if its manipulation leads to any therapeutic benefit. Instead, the muscle expression of DUX4 is causal for the pathology. Notably, to be toxic a constitutive DUX4 expression is required (76). Hence, treatments reducing DUX4 gene expression bear strong therapeutic relevance for FSHD.
Skeletal muscle is the most abundant tissue in our body, accounting for about 40% of its weight. Since FSHD affects several muscle groups throughout the patient body, an efficient and systemic delivery would be required for treating FSHD patients. Physiologically, DUX4 is transiently expressed during the very early stages of embryonic development, it is epigenetically silenced at the 8-cell stage and subsequently maintained repressed in most somatic tissues throughout life. Most FSHD patients develop the disease in adulthood. In healthy adults, DUX4 expression is confined to testis and thymus (1). Hence, future work is required to understand if a body wide DUX4 silencing in adult individuals leads to significant side effects.
Asymptomatic patients have an intermediate level of chromatin relaxation at the FSHD locus, more similar to clinically affected subjects than healthy controls and express detectable DUX4 levels (67,77). Intriguingly, small DUX4 increases in animal models can cause catastrophic effects (78,79). Hence, even small changes in the epigenetic state of the FSHD locus and a partial reduction of DUX4 expression could lead to therapeutic effects. Accordingly, we found that WDR5 targeting significantly rescues the FSHD gene signature, myogenic differentiation and apoptosis despite reducing DUX4 expression of <50%.
Our results indicate that WDR5 targeting promotes cell survival and myogenic differentiation of FSHD muscle cells, while healthy muscle cells are unaffected. While future in vivo studies are required to further support our findings, the lack of a detrimental effect on healthy muscle cells is relevant considering the perspective of therapeutic agents that will have to be administered in a chronic fashion to FSHD patients. In addition, the available evidence further mitigates the concern of targeting WDR5. For example, WDR5 ablation extends life span in C. elegans, whereas inactivation of H3K4 demethylase shortens life span in D. melanogaster (80,81). Moreover, as discussed above, partial/intermittent DUX4 reduction would likely be sufficient to reach therapeutic benefit. Hence, WDR5 most likely don’t need to be completely inhibited in FSHD. Moreover, WDR5 inhibitors with stronger activity and bioavailability are in the making (82), which should further support the possibility to develop a long-term WDR5 targeting modality with minimal side effects.
All in all, our data support the clinical potential for WDR5 targeting as a possible approach to treat FSHD.
DATA AVAILABILITY
All genomic data have been deposited in the GEO database under accession number GSE210610 (https://www.ncbi.nlm.nih.gov/geo/).The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD036038 and 10.6019/PXD036038 (http://www.proteomexchange.org/).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr R. Tawil and the University of Rochester Fields Center for FSHD for providing the human primary muscle cells from healthy donors and FSHD patients used in this work. We thank Dr C. Emerson and the University of Massachusetts Medical School Senator Paul D. Wellstone Muscular Dystrophy Cooperative Research Center for FSHD for providing the human immortalized FSHD muscle cells used in this work. We thank Dr G. Superti-Furga for the pcDNA/FRT/TO/NLS/BirA/Flag vector and the sequence of human WDR5 cloned into the pDonor221, and Dr H. Chang for providing us the BoxB, GAL4-lN and 5X GAL4-UAS firefly Luciferase vectors.
The MF 20 monoclonal antibody developed by Dr D.A. Fischman was obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at the University of Iowa, Department of Biology, Iowa City, IA 52242.
Notes
Present address: Roberto Giambruno, Institute of Biomedical Technologies, National Research Council, 20090, Segrate, Italy and Center for Genomic Science of IIT@SEMM, Fondazione Istituto Italiano di Tecnologia, Milano, Italy.
Contributor Information
Emanuele Mocciaro, Gene Expression and Muscular Dystrophy Unit, Division of Genetics and Cell Biology, IRCCS San Raffaele Scientific Institute, Milano, Italy.
Roberto Giambruno, Gene Expression and Muscular Dystrophy Unit, Division of Genetics and Cell Biology, IRCCS San Raffaele Scientific Institute, Milano, Italy.
Stefano Micheloni, Gene Expression and Muscular Dystrophy Unit, Division of Genetics and Cell Biology, IRCCS San Raffaele Scientific Institute, Milano, Italy.
Filippo M Cernilogar, Division of Molecular Biology, Biomedical Center (BMC), Faculty of Medicine, Ludwig-Maximilians-University (LMU), Munich, Germany.
Annapaola Andolfo, ProMeFa, Proteomics and Metabolomics Facility, IRCCS San Raffaele Scientific Institute, Milano, Italy.
Cristina Consonni, Gene Expression and Muscular Dystrophy Unit, Division of Genetics and Cell Biology, IRCCS San Raffaele Scientific Institute, Milano, Italy.
Maria Pannese, Gene Expression and Muscular Dystrophy Unit, Division of Genetics and Cell Biology, IRCCS San Raffaele Scientific Institute, Milano, Italy.
Giulia Ferri, Gene Expression and Muscular Dystrophy Unit, Division of Genetics and Cell Biology, IRCCS San Raffaele Scientific Institute, Milano, Italy.
Valeria Runfola, Gene Expression and Muscular Dystrophy Unit, Division of Genetics and Cell Biology, IRCCS San Raffaele Scientific Institute, Milano, Italy.
Gunnar Schotta, Division of Molecular Biology, Biomedical Center (BMC), Faculty of Medicine, Ludwig-Maximilians-University (LMU), Munich, Germany.
Davide Gabellini, Gene Expression and Muscular Dystrophy Unit, Division of Genetics and Cell Biology, IRCCS San Raffaele Scientific Institute, Milano, Italy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Research in D.G.’s laboratory is funded by the Italian Ministry of Health [RF-2018-12366631]; European Joint Programme on Rare Diseases [EJPRD20-191]; Italian Association for Cancer Research [IG 2017-ID. 19919]; National Institutes of Health—National Cancer Institute [1R21CA249378-01]; E.M. was supported by the FSHD Society [FSHD-Fall2019-0688952903] and Marie Skłodowska-Curie Individual Fellowships [DREAM-896947]; R.G. was supported by the co-funded fellowship between Vita-Salute San Raffaele University and the European Union Seventh Framework Marie Curie Program [COFUND-INVEST, GA-2010-267264]; work in the G.S. lab is funded by the European Joint Programme on Rare Diseases [EJPRD20-191]. Funding for open access charge: BIBLIOSAN, which has a Read and Publish agreement with OUP.
Conflict of interest statement. None declared.
REFERENCES
- 1. Mocciaro E., Runfola V., Ghezzi P., Pannese M., Gabellini D.. DUX4 role in normal physiology and in FSHD muscular dystrophy. Cells. 2021; 10:3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Preston M.K., Tawil R., Wang L.H.. Facioscapulohumeral muscular dystrophy. Gene Reviews®. 1993; https://www.ncbi.nlm.nih.gov/books/NBK1443/. [Google Scholar]
- 3. Griggs R.C., Tawil R., McDermott M., Forrester J., Figlewicz D., Weiffenbach B.. Monozygotic twins with facioscapulohumeral dystrophy (FSHD): implications for genotype/phenotype correlation. Muscle Nerve. 1995; 18:S50–S55. [PubMed] [Google Scholar]
- 4. Hsu Y.D., Kao M.C., Shyu W.C., Lin J.C., Huang N.E., Sun H.F., Yang K.D., Tsao W.L.. Application of chromosome 4q35-qter marker (pFR-1) for DNA rearrangement of facioscapulohumeral muscular dystrophy patients in Taiwan. J. Neurol. Sci. 1997; 149:73–79. [DOI] [PubMed] [Google Scholar]
- 5. Tupler R., Barbierato L., Memmi M., Sewry C.A., De Grandis D., Maraschio P., Tiepolo L., Ferlini A.. Identical de novo mutation at the D4F104S1 locus in monozygotic male twins affected by facioscapulohumeral muscular dystrophy (FSHD) with different clinical expression. J. Med. Genet. 1998; 35:778–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sakellariou P., Kekou K., Fryssira H., Sofocleous C., Manta P., Panousopoulou A., Gounaris K., Kanavakis E.. Mutation spectrum and phenotypic manifestation in FSHD Greek patients. Neuromuscul. Disord. 2012; 22:339–349. [DOI] [PubMed] [Google Scholar]
- 7. Greco A., Goossens R., van Engelen B., van der Maarel S.M.. Consequences of epigenetic derepression in facioscapulohumeral muscular dystrophy. Clin. Genet. 2020; 97:799–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wijmenga C., Hewitt J.E., Sandkuijl L.A., Clark L.N., Wright T.J., Dauwerse H.G., Gruter A.M., Hofker M.H., Moerer P., Williamson R.. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet. 1992; 2:26–30. [DOI] [PubMed] [Google Scholar]
- 9. Lemmers R.J.L.F., Tawil R., Petek L.M., Balog J., Block G.J., Santen G.W.E., Amell A.M., van der Vliet P.J., Almomani R., Straasheijm K.R.et al.. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat. Genet. 2012; 44:1370–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van den Boogaard M.L., Lemmers R.J.L.F., Balog J., Wohlgemuth M., Auranen M., Mitsuhashi S., van der Vliet P.J., Straasheijm K.R., van den Akker R.F.P., Kriek M.et al.. Mutations in DNMT3B modify epigenetic repression of the D4Z4 repeat and the penetrance of facioscapulohumeral dystrophy. Am. J. Hum. Genet. 2016; 98:1020–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hamanaka K., Šikrová D., Mitsuhashi S., Masuda H., Sekiguchi Y., Sugiyama A., Shibuya K., Lemmers R.J.L.F., Goossens R., Ogawa M.et al.. Homozygous nonsense variant in LRIF1 associated with facioscapulohumeral muscular dystrophy. Neurology. 2020; 94:e2441–e2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Himeda C.L., Jones P.. The genetics and epigenetics of facioscapulohumeral muscular dystrophy. Annu. Rev. Genomics Hum. Genet. 2019; 20:265–291. [DOI] [PubMed] [Google Scholar]
- 13. Lemmers R.J.L.F., van der Vliet P.J., Klooster R., Sacconi S., Camaño P., Dauwerse J.G., Snider L., Straasheijm K.R., van Ommen G.J., Padberg G.W.et al.. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010; 329:1650–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Whiddon J.L., Langford A.T., Wong C.J., Zhong J.W., Tapscott S.J.. Conservation and innovation in the DUX4-family gene network. Nat. Genet. 2017; 49:935–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. De Iaco A., Planet E., Coluccio A., Verp S., Duc J., Trono D.. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet. 2017; 49:941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hendrickson P.G., Doráis J.A., Grow E.J., Whiddon J.L., Lim W., Wike C.L., Weaver B.D., Pflueger C., Emery B.R., Wilcox A.L.et al.. Conserved roles for murine DUX and human DUX4 in activating cleavage stage genes and MERVL /HERVL retrotransposons. 2017; 49:925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gabellini D., Green M.R., Tupler R.. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell. 2002; 110:339–348. [DOI] [PubMed] [Google Scholar]
- 18. Zeng W., de Greef J.C., Chen Y.-Y., Chien R., Kong X., Gregson H.C., Winokur S.T., Pyle A., Robertson K.D., Schmiesing J.A.et al.. Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD). PLoS Genet. 2009; 5:e1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cabianca D.S., Casa V., Bodega B., Xynos A., Ginelli E., Tanaka Y., Gabellini D.. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in fshd muscular dystrophy. Cell. 2012; 149:819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Casa V., Runfola V., Micheloni S., Aziz A., Dilworth F.J., Gabellini D.. Polycomb repressive complex 1 provides a molecular explanation for repeat copy number dependency in FSHD muscular dystrophy. Hum. Mol. Genet. 2017; 26:753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Campbell A.E., Shadle S.C., Jagannathan S., Lim J.-W., Resnick R., Tawil R., van der Maarel S.M., Tapscott S.J.. NuRD and CAF-1-mediated silencing of the D4Z4 array is modulated by DUX4-induced MBD3L proteins. Elife. 2018; 7:e31023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Haynes P., Bomsztyk K., Miller D.G.. Sporadic DUX4 expression in FSHD myocytes is associated with incomplete repression by the PRC2 complex and gain of H3K9 acetylation on the contracted D4Z4 allele. Epigenetics Chromatin. 2018; 11:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Himeda C.L., Jones T.I., Virbasius C.-M., Zhu L.J., Green M.R., Jones P.L.. Identification of epigenetic regulators of DUX4-fl for targeted therapy of facioscapulohumeral muscular dystrophy. Mol. Ther. 2018; 26:1797–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Williams K., Kong X., Nguyen N.V., Mcgill C., Tawil R., Yokomori K., Mortazavi A.. Muscle group specific transcriptomic and DNA methylation differences related to developmental patterning in FSHD. 2021; bioRxiv doi:30 September 2021, preprint: not peer reviewed 10.1101/2021.09.28.462147. [DOI]
- 25. Lee H.Y., Haurwitz R.E., Apffel A., Zhou K., Smart B., Wenger C.D., Laderman S., Bruhn L., Doudna J.A.. RNA-protein analysis using a conditional CRISPR nuclease. Biochemistry. 2013; 110:5416–5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang K.C., Yang Y.W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., Lajoie B.R., Protacio A., Flynn R.A., Gupta R.A.et al.. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011; 472:120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ferri G., Huichalaf C.H., Caccia R., Gabellini D.. Direct interplay between two candidate genes in FSHD muscular dystrophy. Hum. Mol. Genet. 2015; 24:1256–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wiśniewski J.R., Zougman A., Nagaraj N., Mann M.. Universal sample preparation method for proteome analysis. Nat. Methods. 2009; 6:359–362. [DOI] [PubMed] [Google Scholar]
- 29. Rouskin S., Zubradt M., Washietl S., Kellis M., Weissman J.S.. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014; 505:701–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shadle S.C., Bennett S.R., Wong C.-J., Karreman N.A., Campbell A.E., van der Maarel S.M., Bass B.L., Tapscott S.J.. DUX4-induced bidirectional HSATII satellite repeat transcripts form intranuclear double-stranded RNA foci in human cell models of FSHD. Hum. Mol. Genet. 2019; 28:3997–4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Janjic A., Wange L.E., Bagnoli J.W., Geuder J., Nguyen P., Richter D., Vieth B., Vick B., Jeremias I., Ziegenhain C.et al.. Prime-seq, efficient and powerful bulk RNA sequencing. Genome Biol. 2022; 23:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Parekh S., Ziegenhain C., Vieth B., Enard W., Hellmann I.. zUMIs - a fast and flexible pipeline to process RNA sequencing data with UMIs. Gigascience. 2018; 7:giy059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liao Y., Smyth G.K., Shi W.. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019; 47:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jagannathan S., Ogata Y., Gafken P.R., Tapscott S.J., Bradley R.K.. Quantitative proteomics reveals key roles for post-transcriptional gene regulation in the molecular pathology of facioscapulohumeral muscular dystrophy. Elife. 2019; 8:e41740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sergushichev A.A. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. 2016; bioRxiv doi:20 June 2016, preprint: not peer reviewed 10.1101/060012. [DOI]
- 38. Mi H., Huang X., Muruganujan A., Tang H., Mills C., Kang D., Thomas P.D.. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017; 45:D183–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hulsen T., de Vlieg J., Alkema W.. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008; 9:488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Snider L., Geng L.N., Lemmers R.J.L.F., Kyba M., Ware C.B., Nelson A.M., Tawil R., Filippova G.N., van der Maarel S.M., Tapscott S.J.et al.. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet. 2010; 6:e1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sternberg S.H., Haurwitz R.E., Doudna J.A.. Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. RNA. 2012; 18:661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Roux K.J., Kim D.I., Raida M., Burke B.. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012; 196:801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wysocka J., Swigut T., Milne T.A., Dou Y., Zhang X., Burlingame A.L., Roeder R.G., Brivanlou A.H., Allis C.D.. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005; 121:859–872. [DOI] [PubMed] [Google Scholar]
- 44. Yang Y.W., Flynn R.A., Chen Y., Qu K., Wan B., Wang K.C., Lei M., Chang H.Y.. Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. Elife. 2014; 3:e02046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Greenberg J.R. Ultraviolet light-induced crosslinking of mRNA to proteins. Nucleic Acids Res. 1979; 6:715–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sadowski I., Ma J., Triezenberg S., Ptashne M.. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988; 335:563–564. [DOI] [PubMed] [Google Scholar]
- 47. Rickard A.M., Petek L.M., Miller D.G.. Endogenous DUX4 expression in FSHD myotubes is sufficient to cause cell death and disrupts RNA splicing and cell migration pathways. Hum. Mol. Genet. 2015; 24:5901–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Himeda C.L., Debarnot C., Homma S., Beermann M.L., Miller J.B., Jones P.L., Jones T.I. Myogenic enhancers regulate expression of the facioscapulohumeral muscular dystrophy-associated DUX4 gene. Mol. Cell. Biol. 2014; 34:1942–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Balog J., Thijssen P.E., Shadle S., Straasheijm K.R., van der Vliet P.J., Krom Y.D., van den Boogaard M.L., de Jong A., F Lemmers R.J.L., Tawil R.et al.. Increased DUX4 expression during muscle differentiation correlates with decreased SMCHD1 protein levels at D4Z4. Epigenetics. 2015; 10:1133–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huard J., Labrecque C., Dansereau G., Robitaille L., Tremblay J.P.. Dystrophin expression in myotubes formed by the fusion of normal and dystrophic myoblasts. Muscle Nerve. 1991; 14:178–182. [DOI] [PubMed] [Google Scholar]
- 51. Miranda A.F., Bonilla E., Martucci G., Moraes C.T., Hays A.P., Dimauro S.. Immunocytochemical study of dystrophin in muscle cultures from patients with Duchenne muscular dystrophy and unaffected control patients. Am. J. Pathol. 1988; 132:410–416. [PMC free article] [PubMed] [Google Scholar]
- 52. Wallace L.M., Garwick S.E., Mei W., Belayew A., Coppee F., Ladner K.J., Guttridge D., Yang J., Harper S.Q.. DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo. Ann. Neurol. 2011; 69:540–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Geng L.N., Yao Z., Snider L., Fong A.P., Cech J.N., Young J.M., van der Maarel S.M., Ruzzo W.L., Gentleman R.C., Tawil R.et al.. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev. Cell. 2012; 22:38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Corona E.D., Jacquelin D., Gatica L., Rosa A.L.. Multiple protein domains contribute to nuclear import and cell toxicity of DUX4, a candidate pathogenic protein for facioscapulohumeral muscular dystrophy. PLoS One. 2013; 8:e75614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yao Z., Snider L., Balog J., Lemmers R.J.L.F., Van Der Maarel S.M., Tawil R., Tapscott S.J.. DUX4-induced gene expression is the major molecular signature in FSHD skeletal muscle. Hum. Mol. Genet. 2014; 23:5342–5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Homma S., Beermann M.L., Boyce F.M., Miller J.B. Expression of FSHD-related DUX4-FL alters proteostasis and induces TDP-43 aggregation. Ann. Clin. Transl. Neurol. 2015; 2:151–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jagannathan S., Shadle S.C., Resnick R., Snider L., Tawil R.N., van der Maarel S.M., Bradley R.K., Tapscott S.J.. Model systems of DUX4 expression recapitulate the transcriptional profile of FSHD cells. Hum. Mol. Genet. 2016; 25:4419–4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bosnakovski D., Da Silva M.T., Sunny S.T., Ener E.T., Toso E.A., Yuan C., Cui Z., Walters M.A., Jadhav A., Kyba M.. A novel P300 inhibitor reverses DUX4-mediated global histone H3 hyperacetylation, target gene expression, and cell death. Sci. Adv. 2019; 5:eaaw7781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bosnakovski D., Xu Z., Gang E.J., Galindo C.L., Liu M., Simsek T., Garner H.R., Agha-Mohammadi S., Tassin A., Coppée F.et al.. An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J. 2008; 27:2766–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vanderplanck C., Ansseau E., Charron S., Stricwant N., Tassin A., Laoudj-Chenivesse D., Wilton S.D., Coppée F., Belayew A.. The FSHD atrophic myotube phenotype is caused by DUX4 expression. PLoS One. 2011; 6:e26820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Caron L., Kher D., Lee K.L., McKernan R., Dumevska B., Hidalgo A., Li J., Yang H., Main H., Ferri G.et al.. A human pluripotent stem cell model of facioscapulohumeral muscular dystrophy-affected skeletal muscles. Stem Cells Transl. Med. 2016; 5:1145–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Teveroni E., Pellegrino M., Sacconi S., Calandra P., Cascino I., Farioli-Vecchioli S., Puma A., Garibaldi M., Morosetti R., Tasca G.et al.. Estrogens enhance myoblast differentiation in facioscapulohumeral muscular dystrophy by antagonizing DUX4 activity. J. Clin. Invest. 2017; 127:1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kowaljow V., Marcowycz A., Ansseau E., Conde C.B., Sauvage S., Mattéotti C., Arias C., Corona E.D., Nuñez N.G., Leo O.et al.. The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul. Disord. 2007; 17:611–623. [DOI] [PubMed] [Google Scholar]
- 64. Sandri M., El Meslemani A.H., Sandri C., Schjerling P., Vissing K., Andersen J.L., Rossini K., Carraro U., Angelini C.. Caspase 3 expression correlates with skeletal muscle apoptosis in Duchenne and facioscapulo human muscular dystrophy. A potential target for pharmacological treatment?. J. Neuropathol. Exp. Neurol. 2001; 60:302–312. [DOI] [PubMed] [Google Scholar]
- 65. Block G.J., Narayanan D., Amell A.M., Petek L.M., Davidson K.C., Bird T.D., Tawil R., Moon R.T., Miller D.G.. Wnt/β-catenin signaling suppresses DUX4 expression and prevents apoptosis of FSHD muscle cells. Hum. Mol. Genet. 2013; 22:4661–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Statland J.M., Odrzywolski K.J., Shah B., Henderson D., Fricke A.F., van der Maarel S.M., Tapscott S.J., Tawil R.. Immunohistochemical characterization of facioscapulohumeral muscular dystrophy muscle biopsies. J. Neuromuscul. Dis. 2015; 2:291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jones T.I., Chen J.C.J., Rahimov F., Homma S., Arashiro P., Beermann M.L., King O.D., Miller J.B., Kunkel L.M., Emerson C.P.et al.. Facioscapulohumeral muscular dystrophy family studies of DUX4 expression: evidence for disease modifiers and a quantitative model of pathogenesis. Hum. Mol. Genet. 2012; 21:4419–4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Guarnaccia A.d.P., Tansey W.P. Moonlighting with WDR5: a cellular multitasker. J. Clin. Med. 2018; 7:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Siladi A.J., Wang J., Florian A.C., Thomas L.R., Creighton J.H., Matlock B.K., Flaherty D.K., Lorey S.L., Howard G.C., Fesik S.W.et al.. WIN site inhibition disrupts a subset of WDR5 function. Sci. Rep. 2022; 12:1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Grebien F., Vedadi M., Getlik M., Giambruno R., Grover A., Avellino R., Skucha A., Vittori S., Kuznetsova E., Smil D.et al.. Pharmacological targeting of the Wdr5-MLL interaction in C/EBPα N-terminal leukemia. Nat. Chem. Biol. 2015; 11:571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bryan A.F., Wang J., Howard G.C., Guarnaccia A.D., Woodley C.M., Aho E.R., Rellinger E.J., Matlock B.K., Flaherty D.K., Lorey S.L.et al.. WDR5 is a conserved regulator of protein synthesis gene expression. Nucleic Acids Res. 2020; 48:2924–2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wong C.J., Wang L.H., Friedman S.D., Shaw D., Campbell A.E., Budech C.B., Lewis L.M., Lemmers R.J.F.L., Statland J.M., Van Der Maarel S.M.et al.. Longitudinal measures of RNA expression and disease activity in FSHD muscle biopsies. Hum. Mol. Genet. 2020; 29:1030–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. van den Heuvel A., Lassche S., Mul K., Greco A., San León Granado D., Heerschap A., Küsters B., Tapscott S.J., Voermans N.C., van Engelen B.G.M.et al.. Facioscapulohumeral dystrophy transcriptome signatures correlate with different stages of disease and are marked by different MRI biomarkers. Sci. Rep. 2022; 12:1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bosnakovski D., Toso E.A., Hartweck L.M., Magli A., Lee H.A., Thompson E.R., Dandapat A., Perlingeiro R.C.R., Kyba M.. The DUX4 homeodomains mediate inhibition of myogenesis and are functionally exchangeable with the Pax7 homeodomain. J. Cell Sci. 2017; 130:3685–3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Guarnaccia A.D., Rose K.L., Wang J., Zhao B., Popay T.M., Wang C.E., Guerrazzi K., Hill S., Woodley C.M., Hansen T.J.et al.. Impact of WIN site inhibitor on the WDR5 interactome. Cell Rep. 2021; 34:108636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Resnick R., Wong C.-J., Hamm D.C., Bennett S.R., Skene P.J., Hake S.B., Henikoff S., van der Maarel S.M., Tapscott S.J.. DUX4-induced histone variants H3.X and H3.Y mark DUX4 target genes for expression. Cell Rep. 2019; 29:1812–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jones T.I., King O.D., Himeda C.L., Homma S., Chen J.C.J., Beermann M.L., Yan C., Emerson C.P., Miller J.B., Wagner K.R.et al.. Individual epigenetic status of the pathogenic D4Z4 macrosatellite correlates with disease in facioscapulohumeral muscular dystrophy. Clin. Epigenetics. 2015; 7:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bosnakovski D., Chan S.S.K., Recht O.O., Hartweck L.M., Gustafson C.J., Athman L.L., Lowe D.A., Kyba M.. Muscle pathology from stochastic low level DUX4 expression in an FSHD mouse model. Nat. Commun. 2017; 8:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Jones T., Jones P.L.. A cre-inducible DUX4 transgenic mouse model for investigating facioscapulohumeral muscular dystrophy. PLoS One. 2018; 13:e0192657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Greer E.L., Maures T.J., Hauswirth A.G., Green E.M., Leeman D.S., Maro G.S., Han S., Banko M.R., Gozani O., Brunet A.. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 2010; 466:383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Li L., Greer C., Eisenman R.N., Secombe J.. Essential functions of the histone demethylase lid. PLoS Genet. 2010; 6:e1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Teuscher K.B., Meyers K.M., Wei Q., Mills J.J., Tian J., Alvarado J., Sai J., Van Meveren M., South T.M., Rietz T.A.et al.. Discovery of potent orally bioavailable WD repeat domain 5 (WDR5) inhibitors using a pharmacophore-based optimization. J. Med. Chem. 2022; 65:6287–6312. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All genomic data have been deposited in the GEO database under accession number GSE210610 (https://www.ncbi.nlm.nih.gov/geo/).The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD036038 and 10.6019/PXD036038 (http://www.proteomexchange.org/).