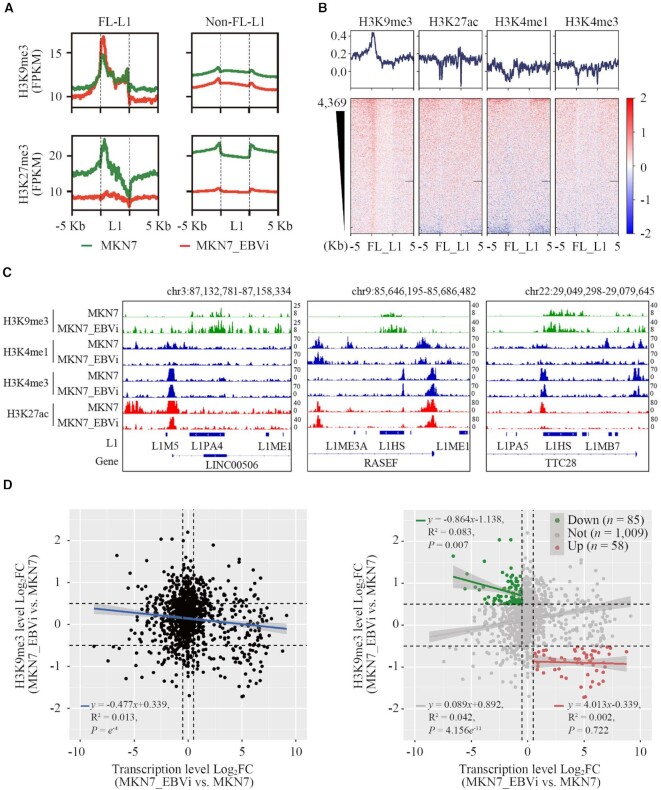
Figure 3.
L1s had aberrant histone modifications in EBVaGC. (A) Average tag density plots showing H3K9me3 and H3K27me3 log2FC enrichment at L1 regions in MKN7_EBVi cells compared with MKN7 cells (MKN7, green; MKN7_EBVi, red; H3K9me3, up; H3K27me3, down; FL-L1, left; non-FL-L1, right). (B) Heatmaps (down) and average tag density plots (up) showing H3K9me3, H3K27ac, H3K4me1 and H3K4me3 log2FC enrichment at FL-L1 (MKN7_EBVi versus MKN7). Black horizon represents the missing signal in the sequencing data. (C) IGV tracks: H3K9me3, H3K4me1, H3K4me3 and H3K27ac signals of three FL-L1P members in MKN7 and MKN7_EBVi cells are shown. The represented signals were normalized by FPKM. Chr, chromosome. (D) The dot plot showing the relationship between log2FC of the FL-L1 transcription level and the FL-L1 H3K9me3 modification level (MKN7_EBVi versus MKN7). Left panel, entire population of FL-L1; right panel, subpopulation analysis.