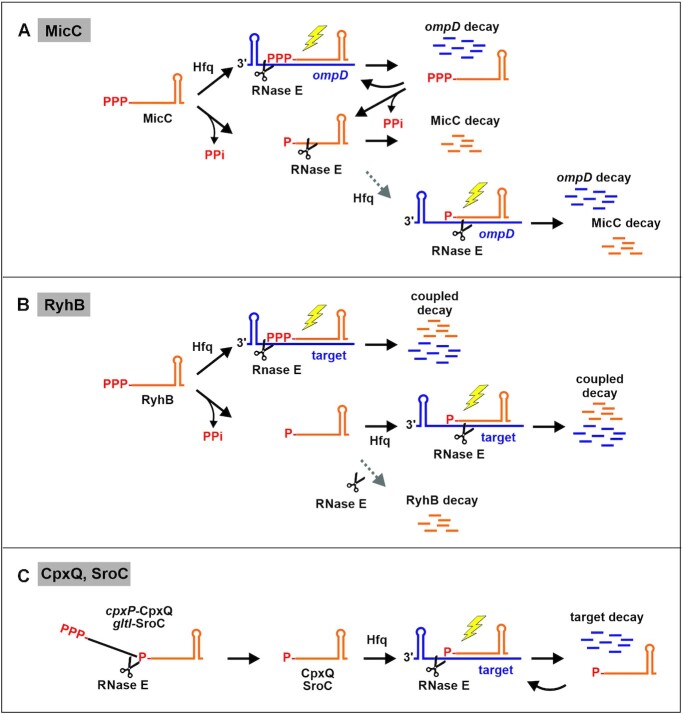
Figure 10.
Model depicting decay pathways and regulatory activities of 5′P- and 5′PPP- sRNA variants as revealed in this study. (A) 5′PPP-MicC is highly stable and regulatory active. It can base pair with its target mRNA ompD and induce RNase E-mediated degradation. These variants may act catalytically until they become dephosphorylated and degraded. In contrast, 5′P-MicC is highly unstable and its major fraction is rapidly degraded before base-pairing can occur (40). Only a minor fraction might be able to find its target to induce mRNA decay (grey dotted arrow). Consequently, MicC displays only a weak regulatory activity in vivo when monophosphorylated. (B) Like MicC, the 5′PPP variant of RyhB is perfectly stable and regulatory active. After base-pairing, RyhB undergoes coupled degradation with its targets (34). The 5′P variant of RyhB is extremely unstable. However, a major fraction of 5′P-RyhB might be able to find its targets and undergoes (coupled) degradation. A minor fraction might be degraded rapidly before base-pairing can occur (grey dotted arrow). (C) sRNAs CpxQ and SroC are produced by RNase E-catalyzed processing from the 3′ UTRs of mRNAs and therefore equipped with 5′P groups. Nonetheless, both sRNAs escape the 5′P-dependent decay pathway of RNase E and are exceptionally stable, regardless of their 5′ phosphorylation state, suggesting that they act in a catalytic manner on their targets. A 5′P for stimulation of target destabilization by RNase E is not required, at least not for sRNAs CpxQ and SroC. A stimulatory effect of the 5′P variants of MicC and RyhB was not observed, but cannot be excluded entirely. sRNA-mediated stimulation of target degradation by RNase E is indicated by a yellow flash.