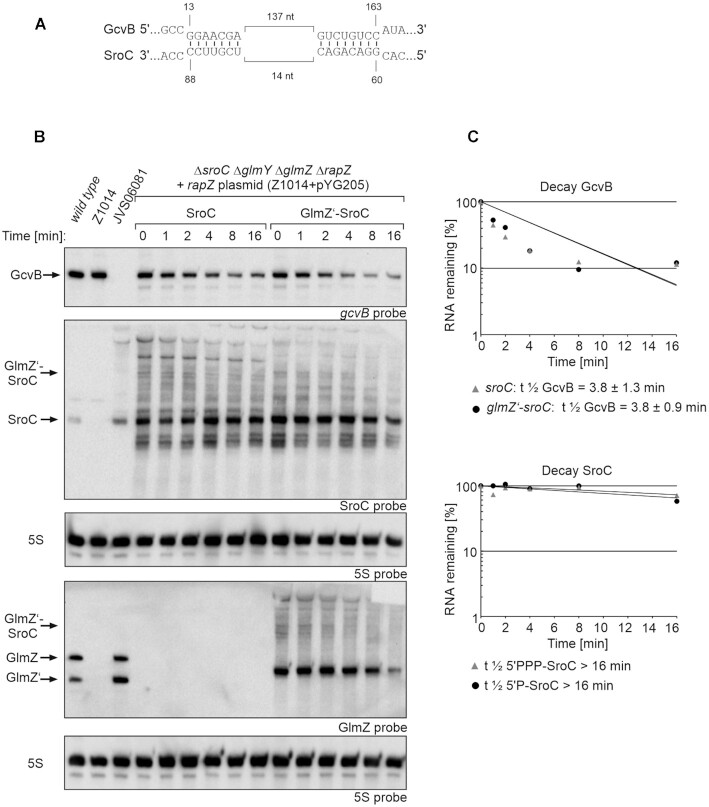
Figure 7.
GcvB decay rates triggered by 5′PPP- and 5′P-SroC and determination of SroC stability. (A) SroC-GcvB base-pairing as shown in Miyakoshi et al. 2015 (60). Base-pairing occurs at two regions that are in close proximity within SroC, but 137 nt apart in GcvB. (B) Northern blot for determination of GcvB decay rates in presence of 5′PPP- SroC and 5′P-SroC and analysis of SroC stability. Rifampicin was added to stop transcription (t0) and samples were removed at indicated times for RNA extraction and Northern blotting. Strain Z1014 (ΔsroC ΔglmY ΔglmZ ΔrapZ) carried plasmid pYG205 (rapZ) and additionally either plasmid pYG277 (sroC) or plasmid pYG276 (glmZ’-sroC). The empty strains S4197 (wild-type), Z1014 and JVS06081 (ΔgcvB) were included as controls (lanes 1–3). The transformants were grown in presence of 0.01 mM IPTG to induce transcription of sroC and glmZ’ fusion genes. Arabinose (0.2%) was added to trigger RapZ-mediated cleavage of the fusion RNAs. Total RNAs were separated on two denaturing 8% PAA gels (1.5 μg each) and subsequently blotted. One blot was probed for GcvB and SroC (first and second panel from top), while the other blot was probed for GlmZ (fourth panel from top). Blots were re-probed for 5S rRNA to obtain loading controls. (C) Semi-logarithmic plots of GcvB and SroC signal intensities for half-life determination. The data are presented as mean, n = 3. Half-lives values are presented as mean ± SD.