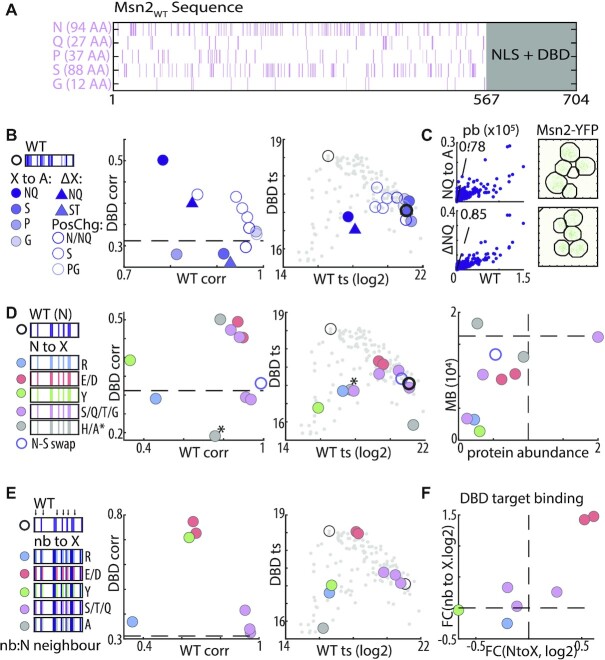
Figure 4.
Asparagine provides the disordered environment required for DNA binding. (A) Distribution of disorder promoting residues within Msn2 nonDBD: shown are the locations and number of the indicated residues within Msn2 IDR. (B, C) Replacing N + Q residues with alanine strongly reduces binding strength and nuclear localization but not binding preferences: binding phenotypes of the indicated mutants are summarized as in Figure 3C (B). Also shown is the similarity of promoter binding preferences between Msn2WT and the N + Q→A and NQ deletion mutant (scatter plots in C, left) and their effect on nuclear localization after EtOH exposure (C, right). (D) Disorder promoting residues retrieve N contribution to IDR-based binding: binding phenotypes of the indicated mutants are summarized as in Figure 3C above (D). Also shown is the absolute motif binding vs. relative protein abundance (right) of the same mutants. Dashed black line indicates Msn2WT to Msn2DBD correlation (left) or Msn2WT abundance and motif binding (right). The NQ to A mutant is indicated with *. (E, F) Acidic residues bias binding towards Msn2DBD promoters: binding phenotypes of the indicated mutants are summarized as in Figure 3C above (E). Change in binding to Msn2DBD targets, measured as fold-change (FC), is also compared between control substitution of N-neighboring and the respective N-to-X substitution (F).