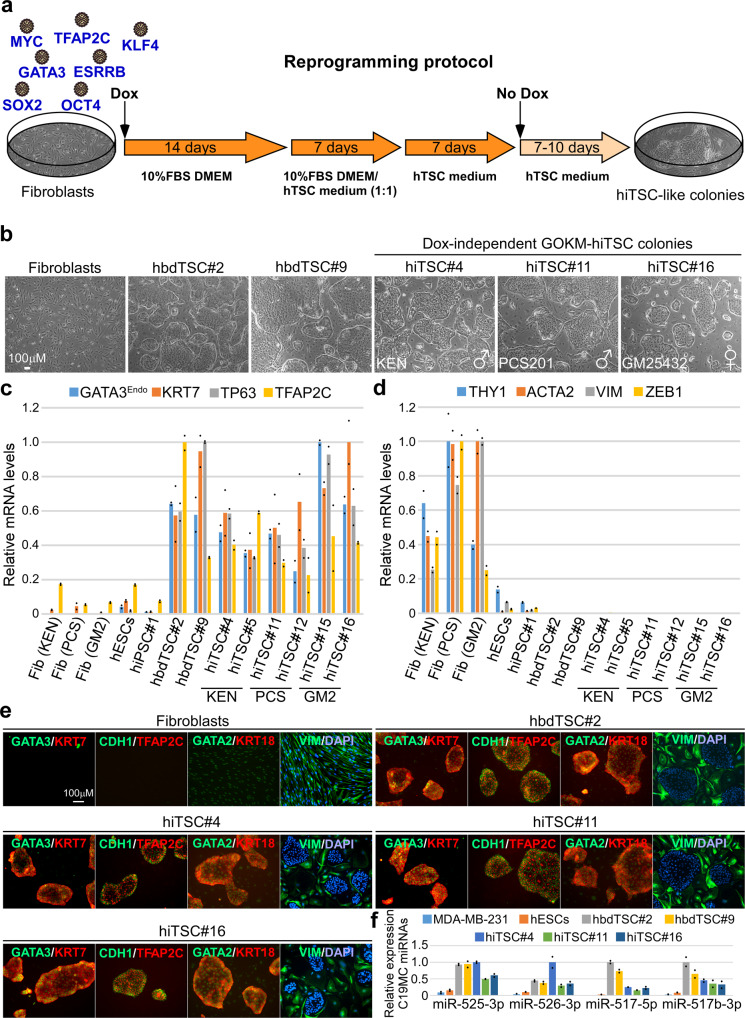
Fig. 1. Ectopic expression of GATA3, OCT4, KLF4 and MYC (GOKM) convert human fibroblasts into trophoblast stem-like cells.
a Schematic representation of the protocol for reprogramming human fibroblasts into human induced trophoblast stem cells (hiTSCs). b Bright field images of human primary fibroblasts, two human blastocyst-derived TSC lines, hbdTSC#2 and hbdTSC#9, and three representative GOKM-derived hiTSC colonies (passage #8–11) originating either from KEN (foreskin fibroblasts, hiTSC#4), PCS201 (foreskin fibroblasts, hiTSC#11) or GM25432 (female adult fibroblasts, hiTSC#16) from three independent reprogramming experiments (n = 3). c, d qPCR analysis of mRNA levels for TSC-specific markers GATA3 (endogenous 5’ UTR expression), KRT7, TP63 and TFAP2C (c) and mesenchymal-specific markers THY1, ACTA2, VIM and ZEB1 (d) in six hiTSC colonies, two hbdTSC lines, three fibroblast lines, hESCs and iPSCs. The indicated hiTSC colonies were derived from three independent reprogramming experiments (n = 3). The highest sample for each gene was set to 1. Results were normalized to the mRNA levels of the housekeeping control gene GAPDH and are shown as fold change. For each sample two replicates were used (n = 2). e Immunofluorescence staining for TSC-specific markers GATA3, KRT7, TFAP2C, GATA2, epithelial markers CDH1 and KRT18, and the mesenchymal marker VIM in parental fibroblasts (KEN) and hbdTSC (hbdTSC#2) controls and in three independent (n = 3) hiTSC clones, hiTSC#4, hiTSC#11 and hiTSC#16. f qPCR analysis for the expression of C19MC miRNA cluster in the indicated samples. hiTSC colonies were derived from three independent reprogramming experiments (n = 3). The highest sample for each miR was set to 1. Results were normalized to the expression levels of the control miR 103a and are shown as fold change. For each sample two replicates were used (n = 2). See also Supplementary Fig. 1. Source data are provided as a Source Data file.