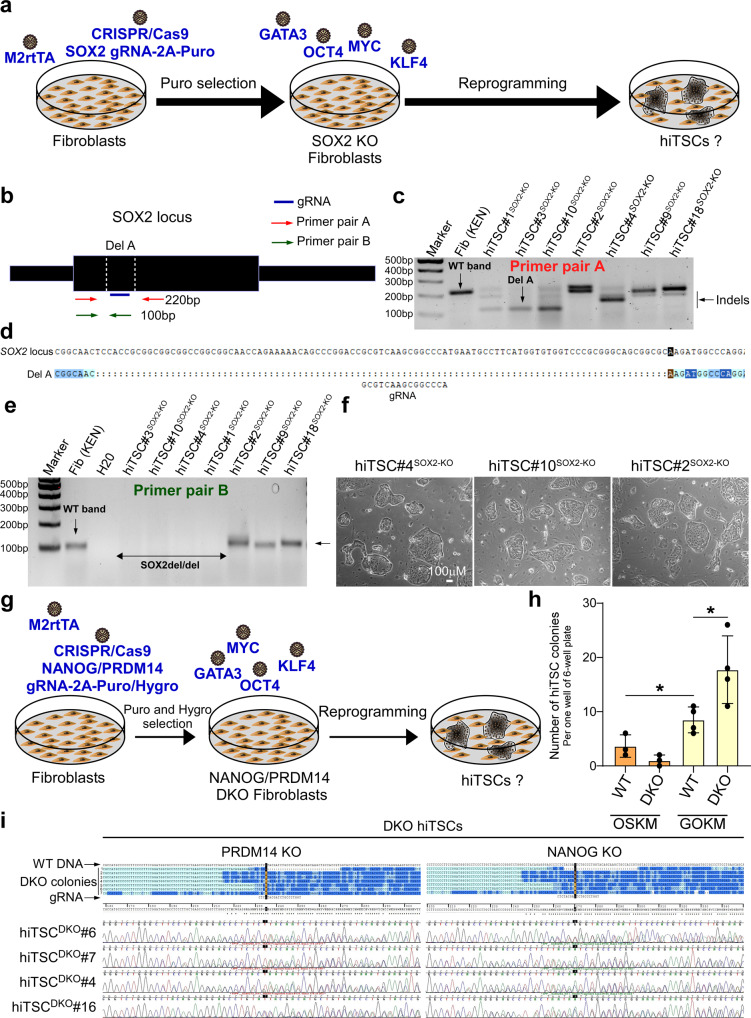
Fig. 7. GOKM produce hiTSCs independently to SOX2 or PRDM14 and NANOG expression.
a Schematic representation for the strategy to generate and reprogram knockout (KO) SOX2 fibroblasts into hiTSCs. b Schematic representation of the SOX2 gene locus, displaying the location of the gRNA used, as well as the primer pairs designed to examine indels. c DNA gel showing a WT band of 219 bp within the SOX2 coding region in WT fibroblasts and the same PCR reaction in seven independent hiTSC clones (n = 7) from SOX2 KO fibroblasts. d Sequence alignment image of one indel event (i.e., Del A) within the SOX2 locus in hiTSC#3SOX2-KO using Sequencher software (demo version). e DNA gel showing a WT band of 100 bp within the SOX2 coding region in WT fibroblasts and the same PCR reaction in seven independent hiTSC clones (n = 7) from SOX2 KO fibroblasts. Note, 4 out of 7 colonies show a bi-allelic deletion within SOX2 coding region (SOX2del/del). f Bright field images of three SOX2 KO hiTSC colonies (n = 3). g Schematic representation for the strategy to double knockout (DKO) NANOG and PRDM14 in fibroblasts and reprogramming into hiTSCs. h Graph depicting the number of hiTSC colonies that emerged either in DKO fibroblasts or in WT controls following reprogramming with GOKM or OSKM. Error bars indicate standard deviation between 3–4 experiments/replicates (for OSKM n = 3 and for GOKM n = 4). * indicates p value of 0.0383 for OSKM WT vs. GOKM WT and 0.0324 for OSKM WT vs. GOKM WT (95% confidence interval of 0.3851–9.282 and 1.081–17.42, respectively), using two-tailed unpaired t-test calculated by GraphPad Prism (8.3.0). Mean values (from left to right) are: 3.667, 1.000, 8.500, 17.75. i Sequences alignment image of various indel events within the NANOG and PRDM14 loci in seven independent hiTSCDKO clones (n = 7) using Sequencher (Demo version). gRNA sequences and Sanger chromatograms are shown for 4 hiTSCDKO clones. Note that a significant enrichment for double KO events is evident in hiTSC clones that were derived from DKO fibroblasts. See also Supplementary Figs. 7–10. Source data are provided as a Source Data file.