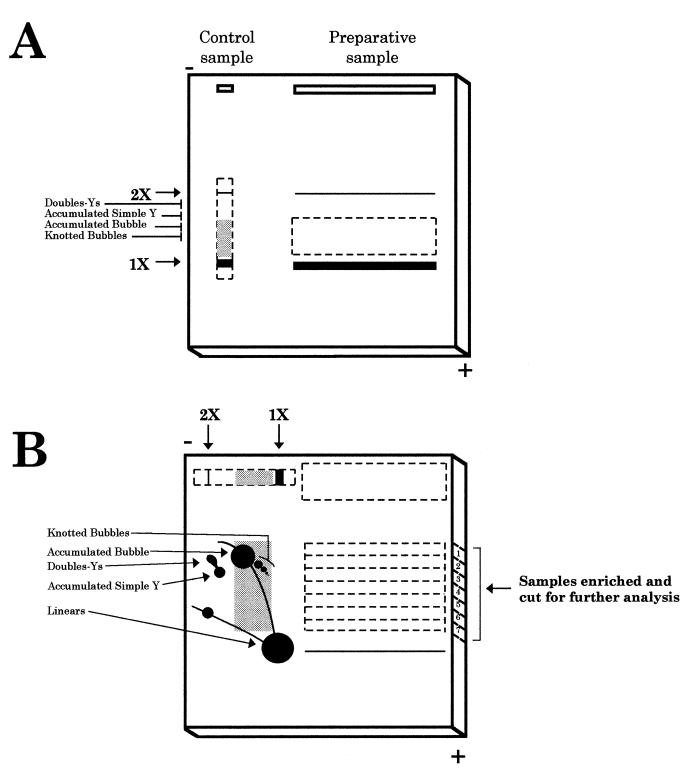
Figure 1.
Schematic representation of the procedure used to isolate specific RIs. (A) First gel electrophoresis. The digested DNA sample (preparative sample) is loaded in a long well. In this way, 200 times the amount of total DNA generally used in a classical 2D agarose gel could be loaded. This would further increase the yield of recovery of specific RIs. An aliquot of this sample was used as a control (control sample). The conditions for the first dimension electrophoresis separate molecules mainly according to their mass. Dotted lines represent the region of the gel cut for the second electrophoresis. Thick lines indicate the migration position of the linear unreplicated molecules. The grey background in the control sample indicates the window selected for isolation of specific RIs from the preparative sample. (B) Second gel electrophoresis. The excised region corresponding to the preparative sample was placed in a new gel that was run in the same orientation as the first electrophoresis. In this way, non-replicated linear molecules, much more abundant than RIs, were selectively eliminated. The lane corresponding to the control sample was excised, rotated 90° and transferred to the same second gel. Low melting point agarose (1%) was then poured. Second electrophoresis conditions emphasised the effect of molecular shape. Horizontal dotted lines in the preparative sample represent the seven regions excised and analysed further. The diagrammatic representation of the control sample indicates the 2D gel pattern for the fragment under study, used as a reference. This procedure allows discrimination between RIs containing different branched structures that co-migrated in the first dimension because they have similar masses.