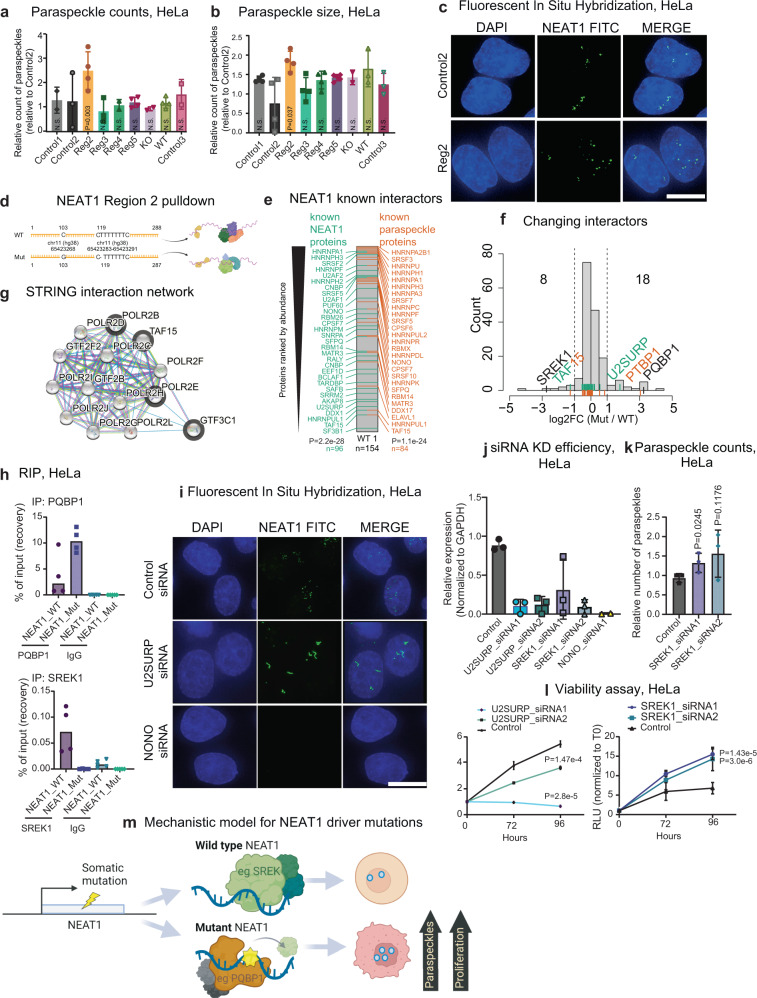
Fig. 6. Mutations at the 5′ end of NEAT1 increase paraspeckle formation and alter its protein interactome.
a Counts of paraspeckles and b paraspeckle size in HeLa cells treated with indicated sgRNAs. Values were obtained from 80 to 100 cells per replicate, with n = 5 (experimental replicates performed at different times). Statistical significance was estimated using one-tailed paired t test. Data show the mean value +/− SD. c Representative images from fluorescence in situ hybridisation (FISH) visualisation of NEAT1 in HeLa cells expressing sgRNAs for Control2 and NEAT1 Region 2. n > 3 biological replicates. Scale bar = 10 µM. d Sequences of biotinylated probes used for the mass-spectrometry analysis of NEAT1-interacting proteins. e Proteins detected by wild-type (WT) NEAT1 probe, filtered for nuclear proteins only, are ranked by intensity and labelled when intersecting previously detected NEAT1-interacting proteins (green) and paraspeckle proteins (orange). Statistical significance was calculated by one-tailed hypergeometric test (to background of nuclear proteins n = 6758). f Histogram shows differential detection of proteins comparing mutated (Mut) and wild-type (WT) probes. Dotted lines indicate log2 fold-change cutoffs of −1/+ 1. g STRING interaction network based on a subset of the proteins lost upon mutation (grey borders) interacting with the RNA polymerase II core complex. h Validations by RNA immunoprecipitation using antibodies for PQBP1 and SREK1. i FISH using NEAT1 probes (green) in HeLa cells treated with indicated siRNAs. Cell nuclei in blue (DAPI). Values were obtained from 80 to 100 cells per replicate, with n = 3 replicates (experimental replicates). Scale bar = 10 µM. j qRT-PCR measurement of RNA levels in HeLa cells after transfection of siRNAs targeting U2SURP and SREK1 genes. Data show the mean value +/− SD of n = 3 independent experiments. k Paraspeckles area in HeLa cells treated with two independent siRNAs targeting SREK1. Measurements from 80 to 100 cells per replicate. Statistical significance was estimated using one-sided paired t test. Data shows the mean value +/− SD of n = 3 replicates (experimental replicates). l Cell viability in siRNA-transfected cells was measured at indicated timepoints. Data shows the mean value +/− SD of n = 3 replicates (experimental replicates). Statistical significance was estimated by using tow-tailed t test. m Proposed model by which somatic mutations in NEAT1 gene can alter protein interactome, increase paraspeckle numbers and boost cell proliferation. Source data are provided as a Source Data file.