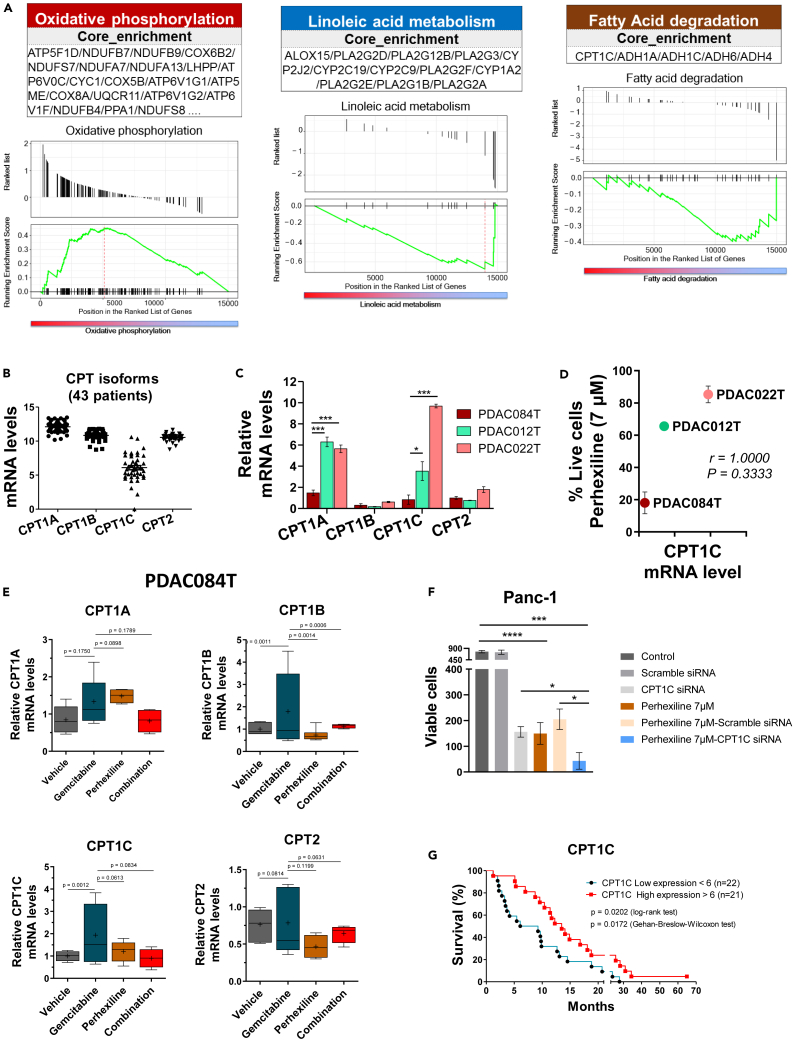
Figure 5.
CPT1C is a key molecular actor behind the response to perhexiline in PDAC
(A) Gene set enrichment analysis showing top upregulated and downregulated pathways in the high responder PDAC cells vs. low responder to FAO inhibition with perhexiline. Oxidative phosphorylation is upregulated, and linoleic acid metabolism and fatty acid degradation are downregulated. Specific pathways are described in each box; CPT1C isoform is found in the downregulated fatty acid degradation pathway.
(B) Gene expression values of CPT isoforms using transcriptomic (RNA-seq) data from 43 primary PDAC cells derived from PDX.
(C) The mRNA levels of CPT1 (A, B, C) and CPT2 isoforms were measured by RT-qPCR in three PDAC cells used in the in vivo experiments. The high responder PDAC084T cells exhibit significantly lower mRNA levels of the CPT1A and CPT1C isoforms compared to the intermediate and low responder PDAC cells (PDAC012T and PDAC022T, respectively). Data are presented as mean ± SEM of three independent experiments performed in duplicates. p values from Student’s t test; ∗p < 0.05, ∗∗∗p < 0.001.
(D) CPT1C mRNA levels correlate with response to perhexiline treatment. The plot shows the mean of CPT1C mRNA levels in the previous panel C, compared with the relative cell viability with perhexiline at 7 μM from Figure 2A. Spearman’s correlation coefficient was used.
(E) CPT mRNA levels in PDAC084T cells were measured by RT-qPCR after 24 h treatment with gemcitabine (1 μM), perhexiline (5 μM), or the combination. Higher mRNA levels of CPT1B and CPT1C are observed in the gemcitabine-treated cells in comparison with the other treatments. Moreover, such increase in gene expression upon chemotherapy seems to be counteracted by perhexiline treatment. Data are presented as mean ± SEM of three independent experiments performed in duplicates. p values shown were calculated from the F-test of equality of variances.
(F) Genetic downregulation of CPT1C by transient transfection of CPT1C siRNA or scramble siRNA (negative control) in the PDAC Panc-1 cell line. Perhexiline treatment was started 24 h after the transfection when CPT1C downexpression was observed (Figure S7), and viability was evaluated by counting the cells 72 h later. Data are presented as mean ± SEM of triplicates in two independent experiments. p values from unpaired Student’s t test; ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(G) Kaplan-Meier survival curves using transcriptomic data from 43 primary PDAC cells derived from PDX.18 Patients are divided into high and low CPT1C gene expression groups (n = 21 and n = 22, respectively; the cutpoint was 6). p values from the log rank test and Wilcoxon test.