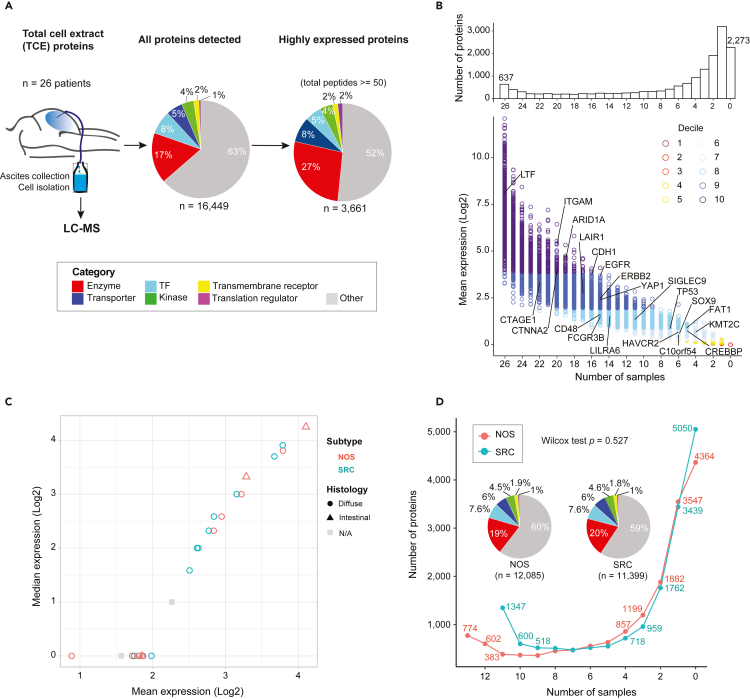
Figure 1.
A landscape of the mass spectrometric-derived cell from 26 advanced gastric cancer patients with peritoneal metastases
From each patient, the ascites was collected and from which the cells were isolated and processed for liquid chromatography-mass spectrometry (LC-MS).
(A) A schematic overview of the protein repertoires from total cell extract (TCE). The total number of proteins detected (total peptides >0) by each approach, the highly expressed proteins (total peptides ≥50) and the proportion of proteins unique to each approach or detected by TCE. The protein categories are annotated in the pie charts based on their molecular functions. TF, transcription factor.
(B) Distribution of protein quantitation measured as mean intensity (Log2 scaled) by the number of samples they are detected in, color coded by the decile. The bar plots on the top display the total counts of proteins quantified across various number of samples. The value 0 stands for the number of proteins that were not detected in any of the samples analyzed. Some representative genes are labeled on the plots.
(C) Scatterplot shows the expression distribution with mean and median values between adenocarcinoma subtypes and histology classifications.
(D) Cumulative number of proteins identified as a function of the patient numbers. Proteins family was shown with pipe plots between NOS and SRC. Statistical method: Wilcox test.