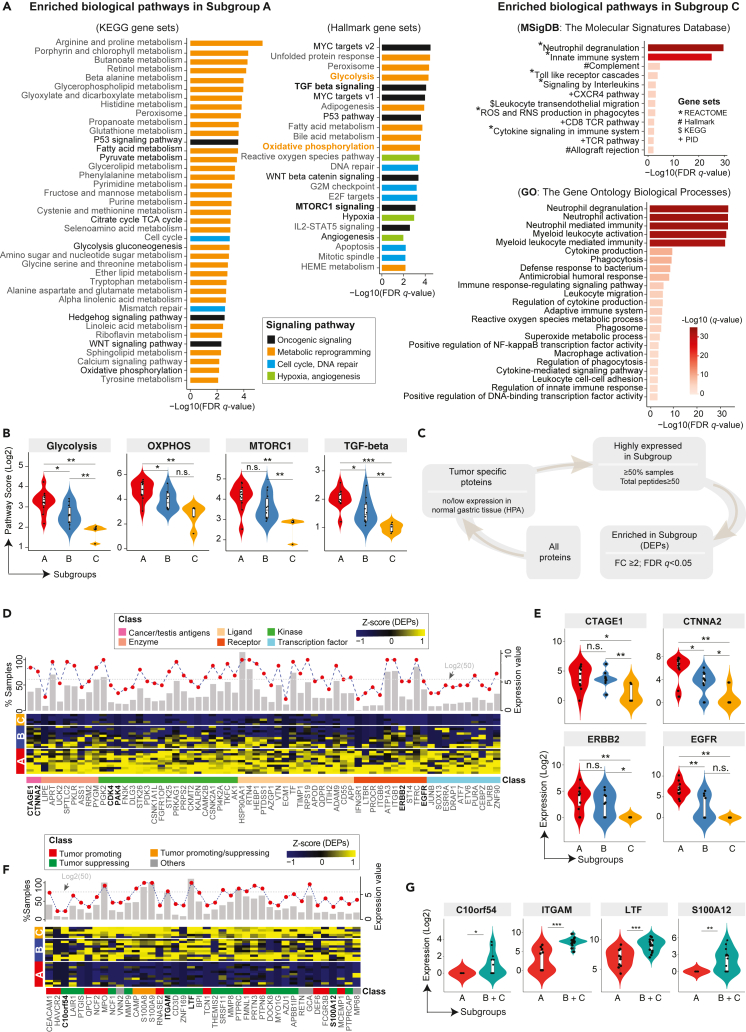
Figure 3.
Enriched TCE proteins and associated biological pathways among three clusters and potential targets
(A) Pathway enrichment analysis identified biological pathways enriched in the subgroup A and C. The curated gene sets were downloaded from the Molecular Signature Database (MSigDB). Both the cancer KEGG (left), Hallmark (middle) and GO (right) gene sets are shown. The pathways are colored by their biological functions. FDR q-value, the p value adjusted for the false discovery rate (FDR). A q-value threshold of 0.01 (1% FDR) is applied.
(B) Violin plots of representative pathways from the subgroup A.
(C) Schematic flow chart showing the filtering process of identifying tumor-specific proteins enriched in the subgroups. The filtering conditions were shown during each filtering step.
(D) A heatmap of highly expressed proteins that are enriched in the Subgroup A from the TCE approach. A list of 65 (out of 560) proteins with known functions are shown. The expression values are Z score transformed. Subgroup classification is annotated on the left, sample names are labeled on the right, and protein names are shown at the bottom, with an annotation track color coded by the protein functional category. Top histogram shows for each listed protein, the fraction of samples with detected protein expression by the TCE approach (n = 26 samples). The red filled circle indicates the mean expression level of each protein averaged across 26 samples (y axis on the right, Log2 transformed). The gray dotted horizontal line annotated the expression level of 50 total peptides.
(E) Violin plots of representative proteins from the panels D. The Mann-Whitney U test was used to calculated the p values for data displayed in panels B and E. ∗∗p < 0.01; ∗p < 0.05; n.s., not statistically significant.
(F) Heatmap of highly expressed proteins that were enriched in the subgroup C from the TCE approach. Proteins that were highly expressed and significantly enriched in the subgroup C are shown at the bottom, with an annotation track color coded by the protein functional category. The expression values are Z score transformed. Subgroup classification is annotated on the left, sample names are labeled on the right, and protein names are shown at the bottom. Top histogram each lists protein, the fraction of samples with detected protein expression by TCE (n = 26 samples). The red filled circles indicate the mean expression level of each protein averaged across all samples (y axis on the right, Log2 transformed). The gray dotted horizontal lines annotate the expression levels of 50 total peptides.
(G) Violin plots of representative proteins from the panels C. The Mann-Whitney U test was used to calculated the p values. ∗∗p < 0.01; ∗p < 0.05; n.s., not statistically significant.