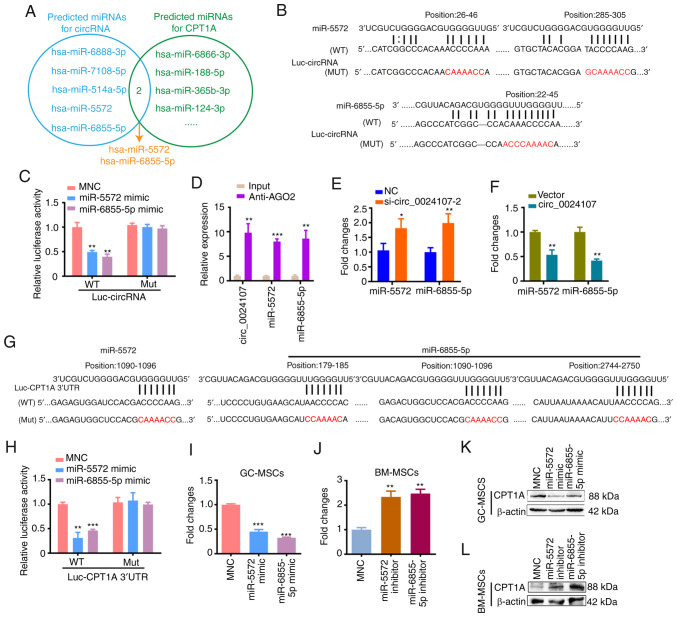
Figure 4.
circ_0024107 upregulates CPT1A expression by sponging miR-5572 and miR-6855-5p. (A) Overlapped miRNAs analysis between the predicted downstream miRNAs sponging by circ_0024107 and the potential miRNAs target regulating CPT1A, predicted using miRanda and TargetScan. (B) circ_0024107-WT and circ_0024107-Mut luciferase reporter construction based on the predicted miRNA binding sites. (C) Luciferase activity assay following the co-transfection of luciferase reporters with miRNA mimics. (D) RIP assay was performed using the anti-AGO2 antibody in GC-MSCs. (E and F) RT-qPCR of the two miRNA levels in (E) circ_0024107-silenced GC-MSCs and (F) circ_0024107-overexpressing BM-MSCs. (G) CPT1A 3′UTR-WT and CPT1A 3′UTR-Mut luciferase reporter construction based on the predicted miRNA binding sites. (H) Luciferase activity assay. (I-L) RT-qPCR and western blot analysis of CPT1A levels in (I and K) miRNA mimic-transfected GC-MSCs and (J and L) inhibitor-transfected BM-MSCs. Values are presented as the mean ± SD (n=3). *P<0.05, **P<0.01 and ***P<0.001, vs. respective control. circRNA, circular RNA; GC-MSCs, gastric cancer-derived mesenchymal stem cells; BM-MSCs, bone marrow-derived mesenchymal stem cells; RT-qPCR, reverse transcription-quantitative PCR; CPT1A, carnitine palmitoyltransferase 1A.