Figure 4.
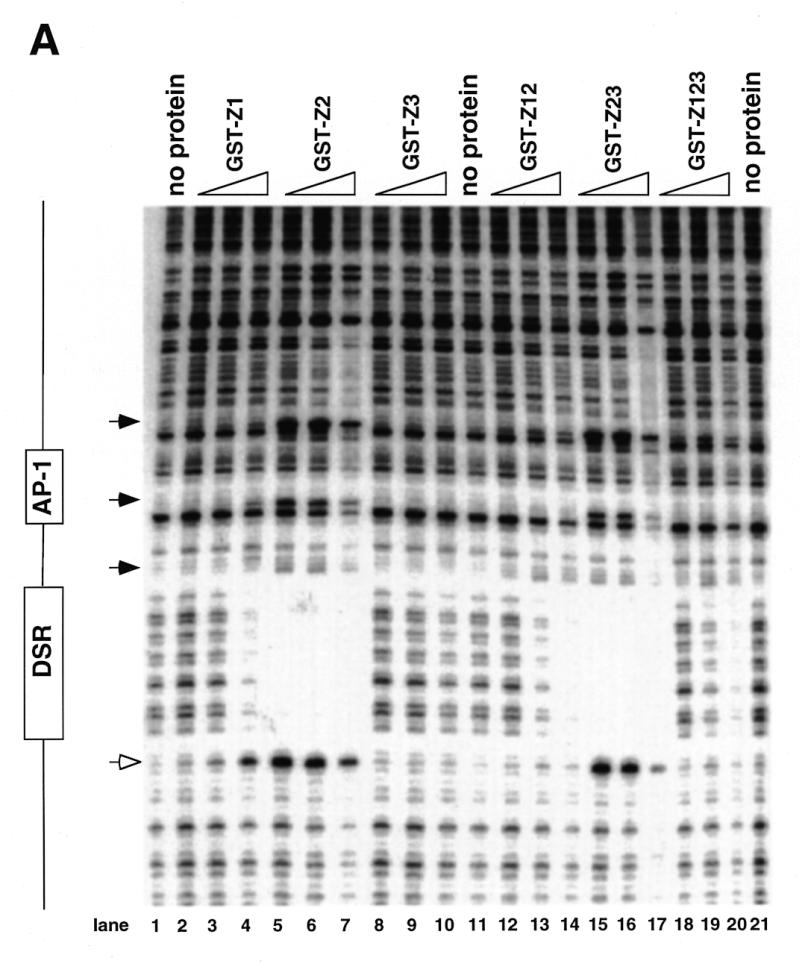
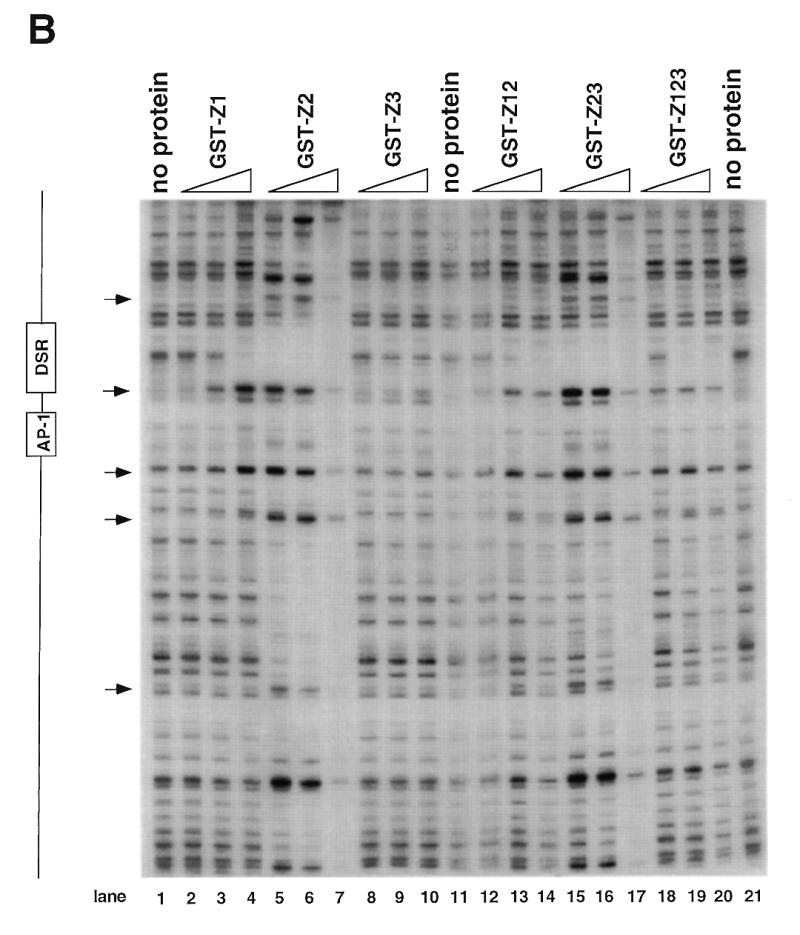
DNase I footprinting of RIP60 on the DHFR-E bent DNA fragment. The indicated RIP60 GST fusion proteins were incubated with end labeled DHFR-E probes and digested with DNase I as described in the Materials and Methods. The digestion products were resolved on sequencing gels and the footprinting patterns were visualized by autoradiography. (A) Nuclease protection patterns for the top strand of DHFR-E. Hypersensitive sites are indicated by arrows. The open arrow indicates a prominent hypersensitive site observed with RIP60 purified from cell extract and by in vivo genomic footprinting of the bent DNA motif in CHO cells (not shown). (B) Nuclease protection patterns for the bottom strand of DHFR-E. Hypersensitive sites are indicated by arrows.
