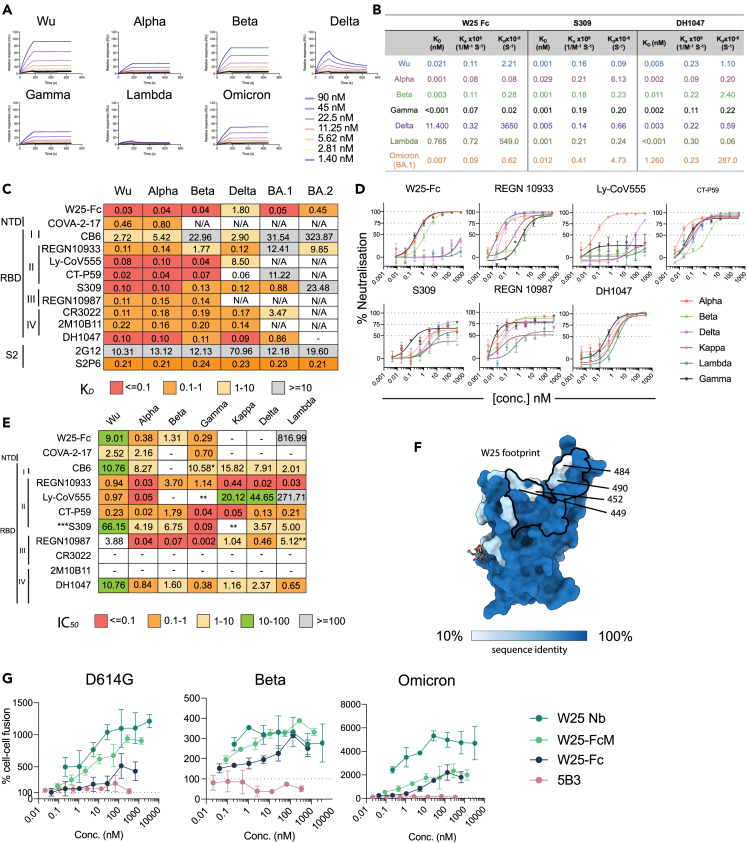
Figure 3.
Broad reactivity of W25-Fc is conferred by conserved patch on SARS-CoV-2 RBD and potential inhibition mechanism of W25
(A) SPR sensograms of W25-Fc. W25-Fc was immobilized on SPR protein A chips. Various concentrations of SARS-CoV-2 spike variant proteins as indicated were injected for 30 s, at 30 μL/min followed by dissociation for 600 s. Dissociation constants (KD) were determined on the basis of fits, applying a 1:1 interaction model. Similar experiments were conducted for S309, DH1047, and C05 (as control, Figure S6).
(B) Summary table showing KD, Ka,Kd of indicated Nb/mAbs.
(C) Kd values from ELISA binding curves of W25-Fc, EUA mAbs, and other epitope-specific mAbs to SARS-CoV-2 spikes variants.
(D) Neutralization curves comparing the sensitivity of live SARS-CoV-2 viruses including Alpha, Beta, Delta, Kappa, Lambda, and Gamma to W25-Fc, EUA, and other RBD-specific mAbs as indicated. The data were analyzed and plotted using nonlinear regression (curve fit, three parameters), and IC50 value was calculated from neutralization curves by Graphpad Prism 8 software.
(E) Summary of IC50 values (nM) of neutralistion of SARS-CoV-2 variants performed in Vero E6 cells. Values that approached 50% neutralization were estimated from (D). Data are generated from two independent experiments, each performed in technical duplicate.
(F) Molecular surface conservation of RBD VOCs. (G) W25 and its derivatives enhance spike-mediated cell fusion. Cell-cell fusion assay was performed. Stable cells expressing the rLuc-GFP components (effector cells) were transiently transfected SARS-CoV-2 spike proteins bearing spike mutations for D614G (top left), Beta (top right), Omicron (Bottom left), or control plasmid (no envelope control). W25 Nb, W25 Dimeric Fc (W25-Fc), or W25 monomeric Fc (W25-FcM) were incubated with effector cells for 1 h prior to co-culturing with target cells (huACE2-HEK293T cells). After 24–48 h, Renilla luciferase was read and analyzed by subtraction with control plasmid treatment. Data are generated from at least three biological replicates. Data are represented as mean ± SEM.