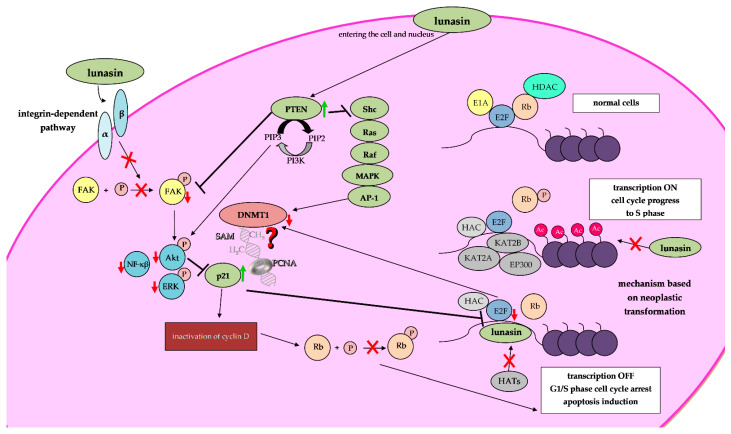
Figure 2.
The cell-specific mechanisms of lunasin activity. The proposed potential repressive effects of lunasin, a potent epinutrient, on DNMT1 transcription and/or DNMT1 activity in cancer cells (middle part of the scheme). Implications of PTEN-mediated negative regulation of intracellular oncogenic signaling pathways, including PI3K/AKT and MAPK/AP-1. PTEN and p21 proteins are negative regulators of AP-1 and E2F, respectively. Those transcription factors (AP-1 and E2F) activate DNMT1 expression due to the presence of binding sites in the DNMT1-regulatory region. Competition of p21 with DNMT1 for the same binding site on PCNA (proliferating cell nuclear antigen). The figure depicts a proposed mechanism of lunasin anti-cancer activity based on the inhibition of integrin signaling and regulation of FAK/AKT/ERK and NF-κB signaling pathways (left part of the scheme) and another mechanism based on an E1A-Rb-HDAC neoplastic transformation model (right part of the scheme). In normal cells in the early G1 phase of the cell cycle (right top part of the scheme), Rb-E2F complex recruits HDACs to maintain the core histones in the repressed/deacetylated state. In the cells being transformed (right middle part of the scheme), during the late G1 phase, E1A (the viral oncogene) inactivates Rb by phosphorylation (Rb-P) and dissociates the Rb-E2F complex, exposing the deacetylated histones to the HATs (i.a., KAT2A, KAT2B, EP300); histone acetylation allows the expression of genes encoding proteins required for S phase and activation of cell cycle progression. In established cancer cell lines (right middle part of the scheme), in which transformation has occurred in the absence of lunasin, HATs acetylated the core histones, turning on cell cycle transcription factors and keeping the acetylated core histones inaccessible and unable to react with added lunasin. Lunasin competes with HATs (i.a., KAT2A, KAT2B, EP300) in binding to hypoacetylated histones (right bottom part of the scheme), repressing transcription that is recognized as abnormal by cells, leading to G1/S phase cell cycle arrest and apoptosis induction. Protein (e.g., Rb) and Protein-P (Rb-P) represent the unphosphorylated and the phosphorylated forms of the protein, respectively. Downregulation (a red down arrow); upregulation (a green up arrow); inhibition (a red X); lunasin as potential modulator of DNA methylation (question mark). Shc, SH2-containing collagen-related proteins; PIP2, phosphatidylinositol (4,5) bisphosphate; PIP3, phosphatidylinositol (3,4,5) trisphosphate; DNMT1, DNA methyltransferase 1; PTEN, phosphatase and tensin homologue; p21 (CDKN1A), cyclin-dependent kinase inhibitor 1A.