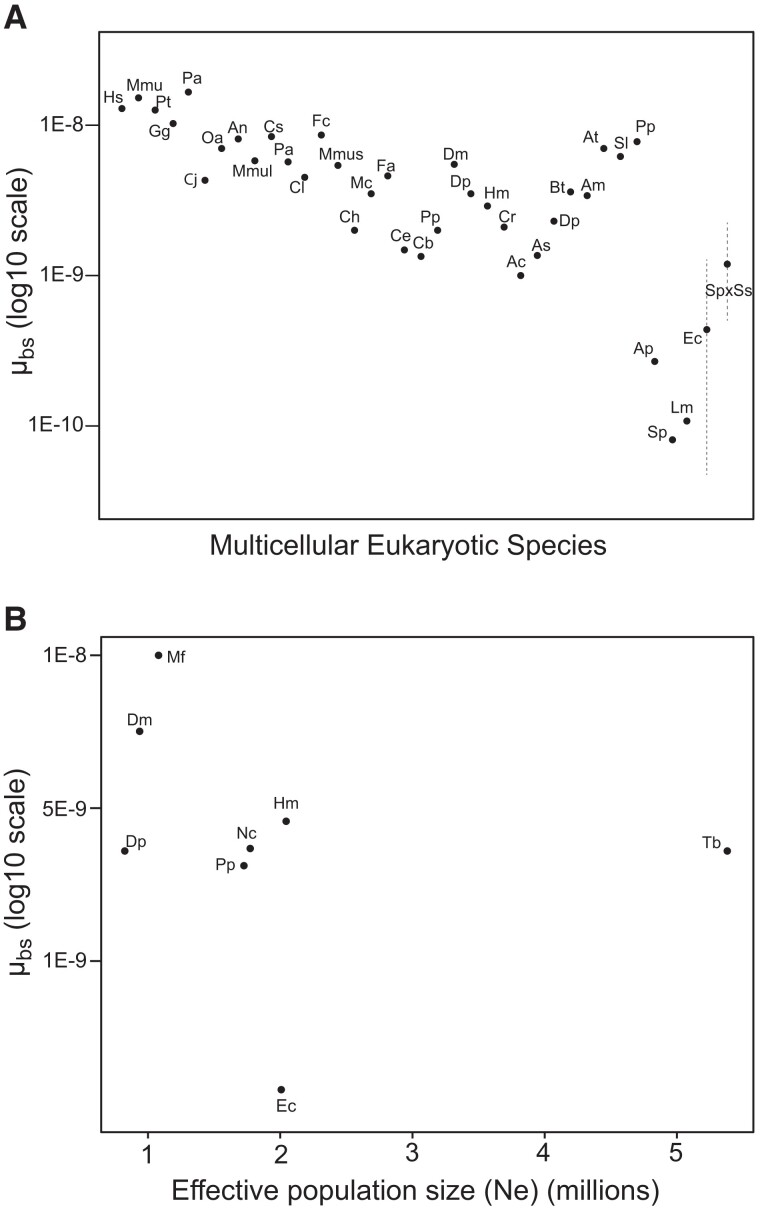
Fig. 1.
Nuclear nucleotide mutation rates µbs of multicellular species (see also supplementary table S1, Supplementary Material online). (A) Species left to right: Homo sapiens, Microcebus murinus, Pan troglodytes, Gorilla gorilla, Pongo abeli, Callithrix jacchus, Ornithorhynchus anatinus, Aotus nancymaae, Macaca mulatta, Chlorocebus sabaeus, Papio anubis, Canis lupus, Felis catus, Mus musculus, Clupea harengus, Malawi cichlids, Ficedula albicollis, C. elegans, Caenorhabditis briggsae, P. pacificus, D. melanogaster, Drosophila pseudoobscura, H. melpomene, Chironomus riparius, Anopheles coluzzii, Anopheles stephensi, D. pulex, Bombus terrestris, Apis mellifera, Arabidopsis thaliana, Silene latifolia, Prunus persica, A. pisum, Spirodela polyrhiza, L. minor, Ectocarpus sp.7, and Scytosiphon hybrid cross. Confidence intervals (CI) are represented for Ectocarpus (Ec) and Scytosiphon hybrid cross (SpxSs). (B) Nuclear nucleotide mutation rates µbs of species with effective population size of the same order as Ectocarpus, between 1 and 5 million (P. pacificus, D. melanogaster, H. melpomene, D. pulex, N. crassa, M. florum, and T. brucei). Effective population size from Lynch et al. 2016.