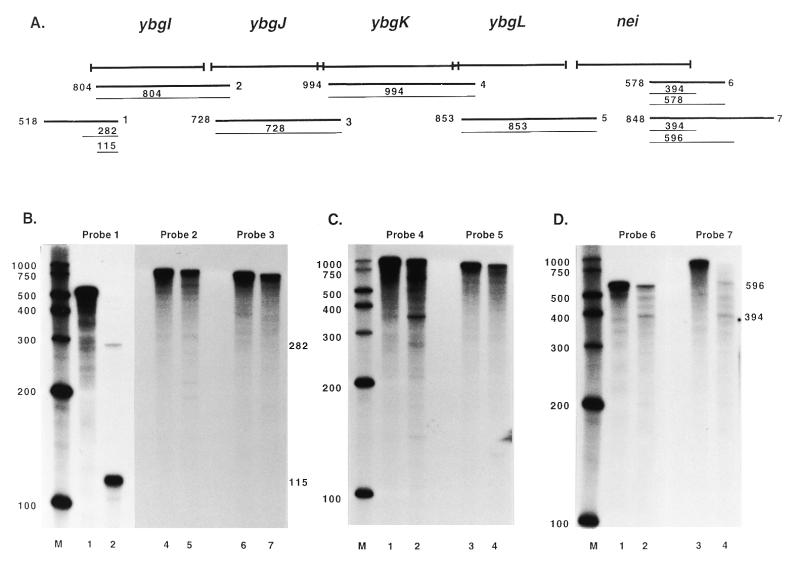
Figure 2.
Mapping of the nei transcript by RPAs. (A) The thick lines designated 1–7 indicate the approximate annealing locations of the probes used in the RPAs. The thin lines indicate the products obtained from RPAs with these probes along with the sizes of the products. The sizes and exact annealing locations of the probes are as follows: probe 1, 518 nt, anneals from 90 bp 3′ to the ybgI start codon to 428 bp 5′ to the ybgI start codon; probe 2, 804 nt, anneals from 146 bp 3′ to the ybgJ start codon to 50 bp 3′ to the ybgI start codon; probe 3, 728 nt, anneals from 87 bp 3′ to the ybgK start codon to 10 bp 3′ to the ybgJ start codon; probe 4, 994 nt, anneals from 81 bp 3′ to the ybgL start codon to 21 bp 3′ to the ybgK start codon; probe 5, 853 nt, anneals from 94 bp 3′ to the nei start codon to 12 bp 3′ to the ybgL start codon; probe 6, 578 nt, anneals from 263 bp 3′ to the nei stop codon to 315 bp 5′ to the nei stop codon; probe 7, 848 nt, anneals from 533 bp 3′ to the nei stop codon to 315 bp 5′ to the nei stop codon. (B–D) The probes used in the RPAs are indicated and lane M contains RNA size markers. Lanes 1, 3 and 5 in (B) and lanes 1 and 3 in (C) and (D) show the full-length probe (probe hybridized with yeast RNA and mock digested; only part of the reaction was loaded so that the signal would not overpower the other lanes). Lanes 2, 4 and 6 in (B) and lanes 2 and 4 in (C) and (D) show RPA results. Numbers beside the panels are in base pairs.