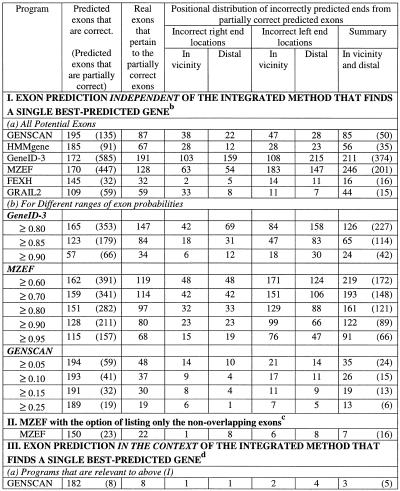
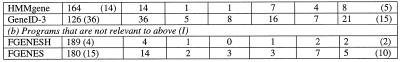
Table 2. Positional characterisation of predicted partially correct exonsa.
aThe test sequences contained 198 exons with both the boundaries as coding.
bThe gene prediction programs used can list all potential splice sites as well as different possible exons. For calculations under ‘independent’, all predicted potential exons (not necessarily from the single best-predicted gene) were considered. MZEF was executed with the option of listing up to 10 overlapping exons. The sub-optimal exons from GENSCAN differ from the potential exons as predicted by other programs in that they are somewhat dependent upon the context. The sub-optimal exon is in-frame in one of the possible valid parses of the sequence. The sub-optimal exons from HMMgene are from the top five best-predicted genes and thus they are dependent upon the context.
cIn the case of MZEF, the predicted exon has a posterior probability value of >0.50. Of a set of predicted overlapping exons, the one with the highest posterior probability is selected as the optimal exon.
dOnly the internal exons from the predicted optimal gene were considered.