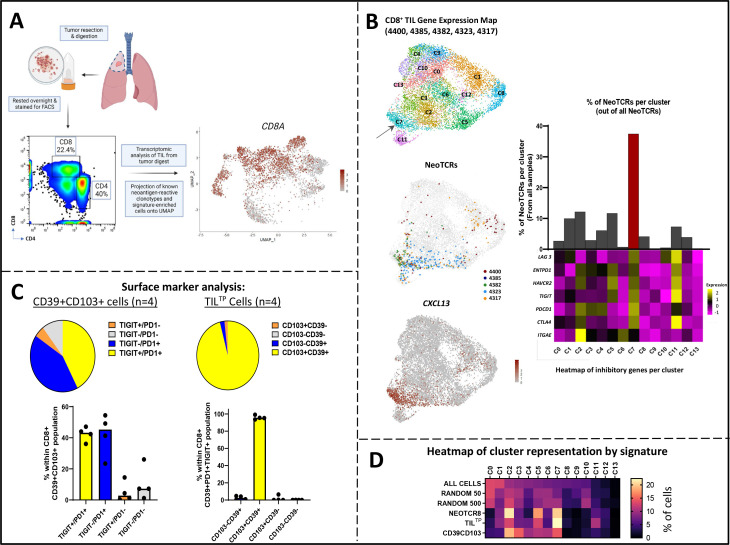
Figure 1.
(A) CD8+ TIL populations were isolated from fresh tumor digest. Single-cell transcriptomic analysis was performed on the patient’s TIL from tumor digest and known neoantigen-reactive clonotypes and signature-enriched cells were projected onto a Uniform Manifold Approximation and Projection (UMAP). (B) Integrated single-cell transcriptomic analysis showing UMAP-based projection of CD8+ TIL states from five patient samples (4400, 4385, 4382, 4323, 4317; left, top); cluster 7 (C7) indicated by arrow. Back-projection of 31 known neoantigen-reactive TCRs (NeoTCR) clones onto UMAP plot (left, middle). UMAP transcriptomic map of CD8+ TIL shows NeoTCR8 transcriptomic state showing co-localization of CXCL13-expressing cells (left, bottom). Bar graph indicating the percentage of known neoantigen-reactive TCRs (NeoTCRs) within each cluster, with the bar for NeoTCR8 state C7 denoted in red (right, top). Heatmap indicating average transcriptomic expression of inhibitory genes within each cluster (right, bottom). (C) Surface marker analysis of CD8+ CD39+CD103+ (double positive) and ‘triple positive’ (CD8+ PD-1+ TIGIT+ CD39+) TILTP cells of four patients (4400, 4385, 4323, 4393). Pie chart of proportion of cells expressing TIGIT and PD-1 within the CD39+CD103+ TIL subset; bar graph with the median of individual patient data (n=4) represented by black dots (left). Pie chart of proportion of cells expressing CD39 and CD103 within the TILTP subset; bar graph with the median of individual patient data represented by black dots (right). (D) Heatmap showing the relative frequency of cells within each cluster from single-cell RNA from figure 1B, that can be identified by the top decile within each signature. ‘All Cells’ includes all cells present in the transcriptomic UMAP. ‘Random50’ and ‘Random500’ represent signatures derived from random genes sets of 50 and 500 genes as negative control signatures, respectively. TCR, T-cell receptor; TIL, tumor-infiltrating lymphocytes.