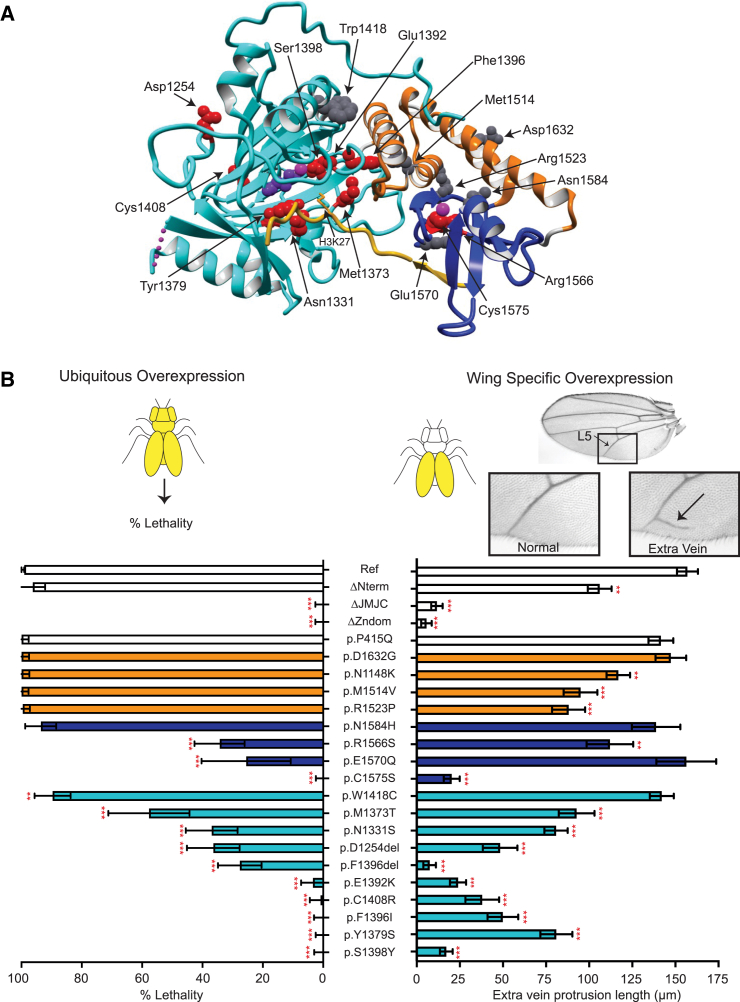
Figure 2.
Analysis of KDM6B PAVs via protein 3D structure analysis and a dual Drosophila gain-of-function assay
(A) KDM6B fragment (PDB: 5OY3, aa 1157–1639) bound to the H3 tail fragment (aa 17–33). The JmJC domain is shown in cyan with 2-oxoglutaric acid (purple) bound with an Fe ion (magenta), which is necessary for the enzymatic demethylation of H3K27. The Zn-containing domain is shown in blue with Zn ion (magenta). Two out of three JmJC and Zn-containing domain-stabilizing linkers are also visible in the structure (orange). The H3 tail with K27 residue positioned into the active center of the JmJC domain is shown in yellow. Amino acids affected by missense or in-frame indels are shown as balls (pathogenic, red; VUSs, gray), affecting all shown domains as well as binding to H3 tail (see Table S3 for more details on specific variants).
(B) A dual Drosophila gain-of-function assay was used to assess the disruptive potential of KDM6B PAVs. Ubiquitous overexpression (left) of KDM6Bref with the UAS/Gal4 system results in complete lethality. Percent lethality assessed for KDM6BΔNterm, KDM6BΔJmJC, KDM6BΔZndom, KDM6BP415Q as a benign control, and 18 KDM6B variants were compared to KDM6Bref (chi-squared test). n = 50–230 flies for each genotype; data are represented as mean ± 95% confidence interval. Wing-specific overexpression (right) of KDM6Bref in the fly wings results in the formation of an extra vein protruding off the L5 vein. The length of the extra vein was compared to KDM6Bref (Dunnet’s test). n = 18–35 flies for each sample; data are represented as mean ± SEM. PAVs are colored on the basis of the domain (JmJC, cyan; Zn-containing, blue; stabilizing linkers, orange; no domain, white; same as in Figures 1B and 2A). ∗∗p < 0.01, ∗∗∗p < 0.0001.