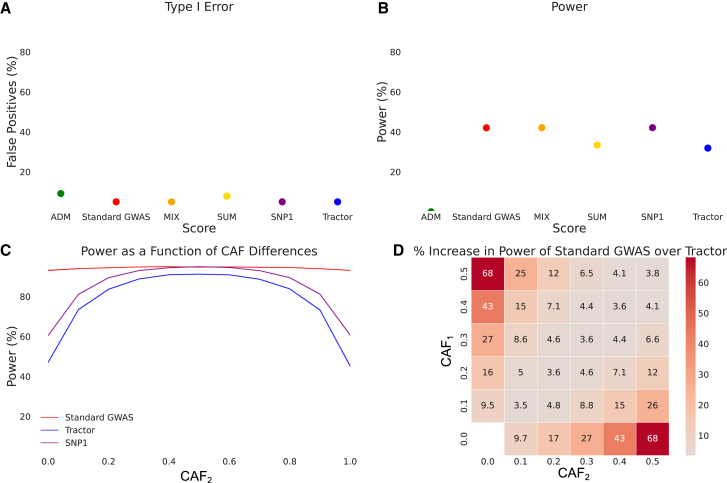
Figure 2.
Association statistics in the absence of HetLanc
(A) Type I error for association statistics. Type I error calculated as the percent of null SNPs with a significant association detected. 95% confidence interval too narrow for display.
(B) Power for association statistics. Power calculated as the percent of simulations to successfully recover the causal variant. Odds ratios OR1 = OR2 = 1.2. 95% confidence interval too narrow for display.
(C) Power for a standard GWAS, SNP1, and Tractor as CAF2 is varied between 0.0 and 1.0 and CAF1 is fixed at 0.5. Power for all three methods varies as CAF difference varies. 95% confidence interval too narrow for display.
(D) Heatmap of percent increase in power of a standard GWAS over Tractor when . Causal allele frequencies CAF1 and CAF2 varied from 0.0 to 0.5 in increments of 0.1.
All simulations are for case-control (A and B) or quantitative (C and D) traits simulated 1,000 times for a population of 10,000 individuals with 100 genotypes each with global ancestry proportion 50/50. Power calculated using (A) nominal threshold p value < 0.05, (B) Bonferroni-corrected threshold p value < 1 × 10−5, or (C and D) standard threshold p value < 5 × 10−8. (A and B) Case-control traits have case-control ratio 1:1, 10% case prevalence, and CAF1 = CAF2 = 0.5. (C and D) Quantitative traits have heritability = 0.005. Heritability, global ancestry, causal effect size , and overall CAF do not qualitatively impact these results (Figures S1–S3).