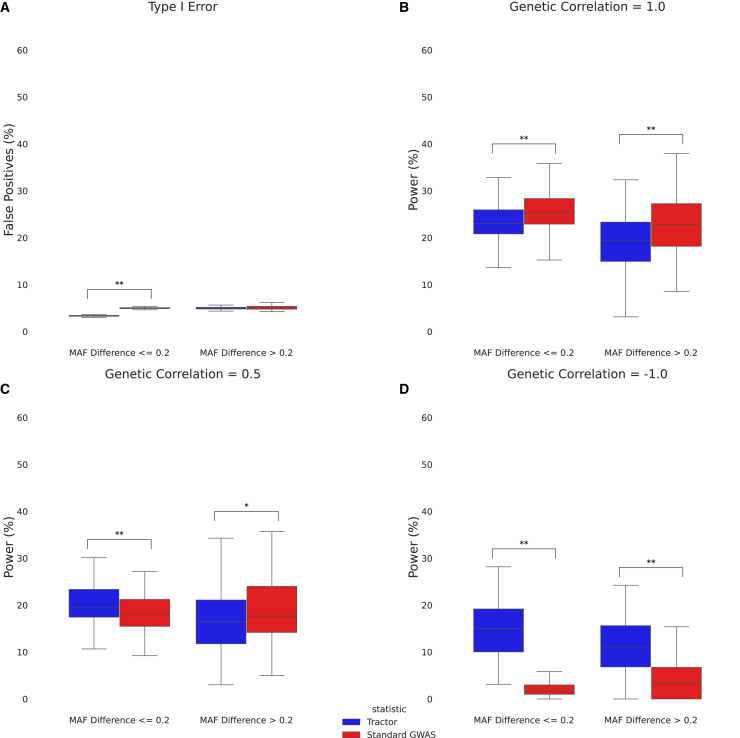
Figure 4.
Effect size heterogeneity in the context of polygenicity
(A) Boxplot of type I error for Tractor and a standard GWAS split by non-differentiated (MAF difference 0.2) and differentiated (MAF difference > 0.2) SNPs.
(B) Boxplot of power for Tractor and a standard GWAS in the case of no effect size heterogeneity split by non-differentiated and differentiated SNPs.
(C) Boxplot of power for Tractor and a standard GWAS in the case of effect size heterogeneity split by non-differentiated and differentiated SNPs.
(D) Boxplot of power for Tractor and a standard GWAS in the case of opposite effect sizes split by non-differentiated and differentiated SNPs.
All simulations used real UKBB admixed genotypes and simulated phenotypes with 100 causal SNPs and a total additive genetic heritability of = 0.5 (see subjects and methods). “∗” indicates a nominally significant p value (<0.05). “∗∗” indicates a Bonferroni-corrected significant p value (<1.28 × 10−3). The boxes show the inter-quartile range while the whiskers show the rest of the distribution (not including outliers).