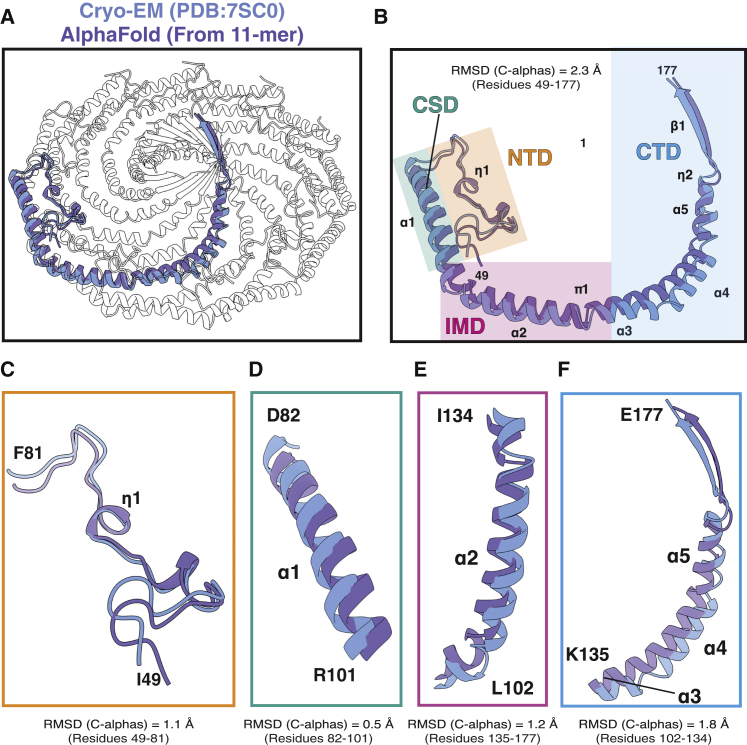
Figure 5.
Comparison of the CAV1 protomers from the cryo-EM structure and 11-meric complex of CAV1 predicted by AF2.2. (A) The CAV1 8S complex (PDB: 7SC0) is shown with a single protomer colored blue and overlayed with an aligned AF2.2 protomer taken from the AF2.2-predicted CAV1 11-mer in purple. (B) Overlay of a single protomer from the cryo-EM structure and the AlphaFold2 CAV1 protomer. The secondary structure assignments for the cryo-EM structure are labeled. The coordinates were aligned by their alpha carbons, and their root-mean-square deviation values are shown. The first 48 residues were not included in the alignments. Each region is highlighted by a different color. Orange, residues 49–81 of the N-terminal domain; teal, SD; pink, intramembrane domain; and blue, residues 135–177 of the C-terminal domain. (C–F) Each of the indicated regions was aligned individually and root-mean-square deviation values calculated. Experimental structures are shown in blue, and predicted structures are shown in purple.