Abstract
Background:
Management of thyroid nodules may be inconsistent by different observers and time consuming for radiologists. An artificial intelligence system based on deep learning may improve radiology workflow for management of thyroid nodules.
Purpose:
To develop a deep learning algorithm that uses thyroid ultrasound images to decide whether a thyroid nodule should undergo a biopsy, and to compare the performance of such algorithm to radiologists following ACR TI-RADS.
Materials and Methods:
In this IRB-approved, HIPAA-compliant study, 1377 thyroid nodules from 1230 patients with complete imaging data were retrospectively analyzed. Their malignancy status was determined by either fine-needle aspiration or surgical histology and used as the gold standard. A radiologist assigned ACR TI-RADS features to each nodule. We trained a multi-task deep neural network to provide biopsy recommendations for thyroid nodules based on two orthogonal ultrasound images as the input. In the training phase, the deep learning algorithm was first evaluated using 10-fold cross-validation and then validated on an independent set of consecutive 99 cases not used for model development. The sensitivity and specificity of our algorithm were compared to (1) a consensus of three ACR TI-RADS committee experts and (2) nine other radiologists, all of whom interpreted thyroid ultrasound in clinical practice.
Results:
On the 99 test cases, the proposed deep learning algorithm achieved 87% sensitivity, the same as expert consensus, and higher than five of nine radiologists. The specificity of the deep learning algorithm was 52% which was similar to ACR TI-RADS committee expert consensus (51%) and higher than seven of nine other radiologists. The mean sensitivity and specificity for the nine radiologists was 83% and 48%, respectively.
Conclusions:
A deep learning algorithm for thyroid nodule biopsy recommendations performed with similar sensitivity and specificity compared to ACR TI-RADS committee expert radiologists using ACR TI-RADS guidelines.
Keywords: Thyroid Nodules, Ultrasound, TI-RADS, Deep learning
SUMMARY STATEMENT
In this paper, we showed that deep learning is a highly promising tool that can be applied in the interpretation of thyroid ultrasound and decision making process for thyroid nodules.
INTRODUCTION
Imaging with ultrasound remains the most accurate method to guide recommendation for management of thyroid nodules (1), although interpretation variability and overdiagnosis represent continual challenges (2)(3). To help radiologists improve consistency, several organizations have developed imaging criteria to aid in the selection of nodules recommended for fine-needle aspiration (FNA) biopsy. In 2017, the American College of Radiology published its Thyroid Imaging, Reporting, and Data System (ACR TI-RADS) (4). Similar to its predecessors, ACR TI-RADS is based on ultrasound features as well as maximum nodule size. ACR TI-RADS has been shown to increase accuracy and specificity compared to other systems (5), enhance report quality, and improve recommendations for management (6).
Despite these potential benefits, certain barriers may prevent radiologists from adopting or using ACR TI-RADS. First, a high interobserver variability among radiologists’ interpretations has been shown when using the system (κ=0.51) (2). Such variability may lead to inconsistent recommendations for nodule management between readers. Second, evaluating multiple nodules (with multiple features per nodule) can be labor intensive and could be more time-consuming for some radiologists. Any practice that adds time to an already busy radiology workflow could serve as a disincentive for adopting best practices.
It is for these types of challenges that the medical community has started turning to deep learning (7). Deep learning represents an approach to artificial intelligence that has been increasingly applied throughout medicine, with emerging applications in fields such as dermatology (8), ophthalmology (9), and particularly radiology (10,11). Recent deep learning research in radiology has shown algorithm performance comparable to radiologists (12), and as the field continues to grow, the variety and number of potential uses for deep learning continue to increase. Some of the challenges with thyroid ultrasound interpretation and reporting data systems such as ACR TI-RADS represent problems that may be solved through deep learning applications.
The aim of our study was to design a deep learning algorithm to interpret ultrasound images of thyroid nodules and to make management decisions. We also aimed to compare the performance of the algorithm to radiologists with varying expertise (3 to 32 years), all of whom interpreted thyroid ultrasound in clinical practice, following ACR TI-RADS interpretation criteria.
MATERIALS AND METHODS
Study population
In this IRB-approved, Health Insurance Portability and Accountability Act compliant study, we retrospectively analyzed a dataset of thyroid nodules. The initial population included 1631 nodules in 1439 adult patients from a single institution who underwent diagnostic thyroid ultrasound examinations and ultrasound guided fine-needle aspiration of a focal thyroid nodule between August 2006 and May 2010. It was refined by excluding 203 nodules in 172 patients with initial non-diagnostic or indeterminate cytology and without subsequent diagnostic cytology or histology. Nodules with incomplete imaging data, specifically cases in which images from one or both orthogonal planes were missing (n=15), were also excluded. In addition, to facilitate nodule detection (based on a method utilizing calipers), cases that did not contain images with proper caliper measurement marks (at least one caliper measurement in one plane and two in the other) were excluded (n=36). This resulted in 1377 nodules from 1230 patients. In the final sets for the analysis, there were 1278 nodules from 1139 patients in the training set and 99 nodules from 91 patients in the test set (Figure 1). The 99 test nodules were not used during algorithm development. They were analyzed by multiple readers in a previous study (details below) (5).
Figure 1:
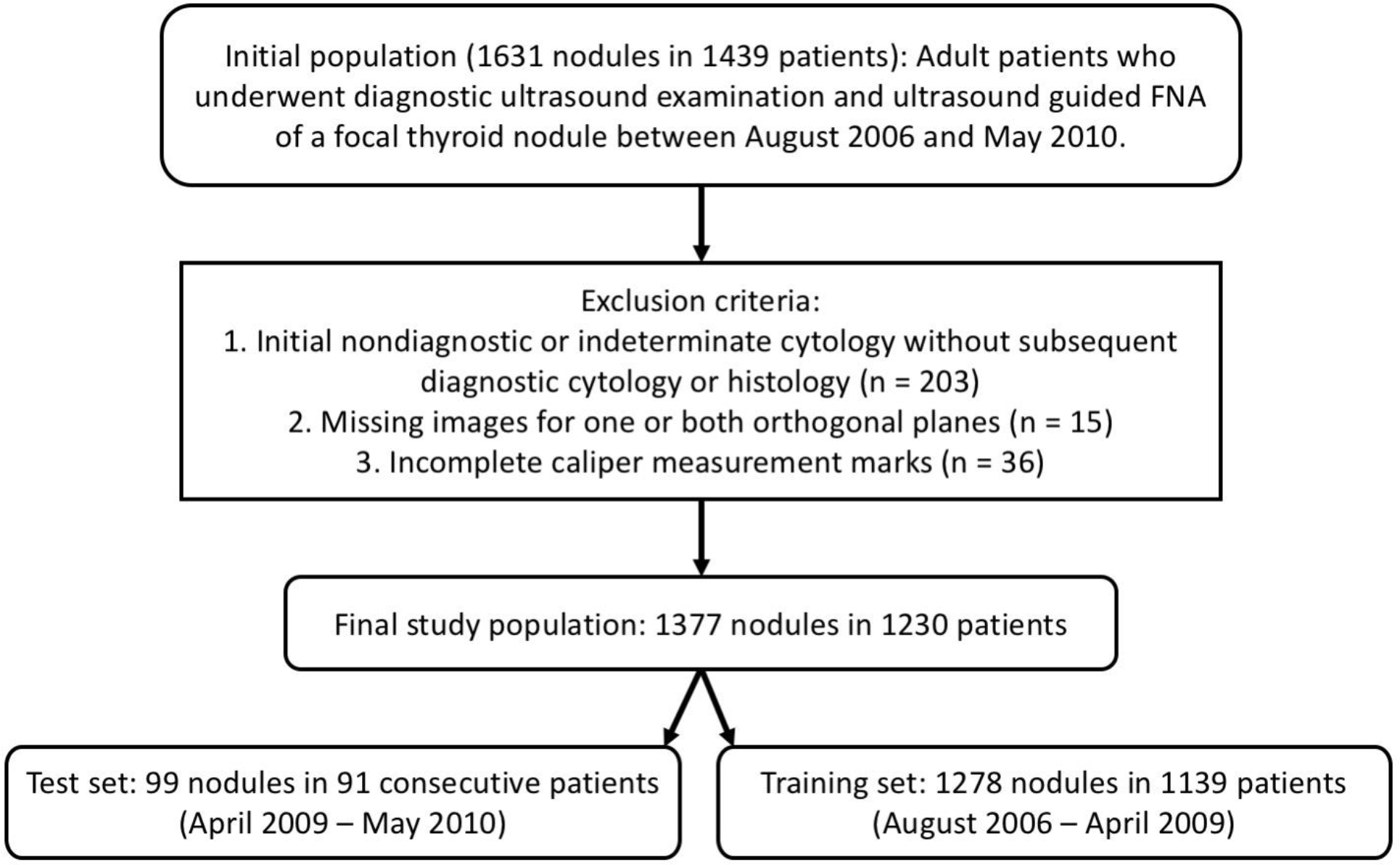
A flowchart showing inclusion criteria for initial population and exclusion criteria for final study population.
The ultrasound examinations were performed using a variety of commercially available units (Antares and Elegra [Siemens, Munich, Germany], ATL HDI 5000 and iU22 [Philips, Amsterdam, Netherlands], and Logic E9 [General Electric, Boston, MA]) equipped with 5–15 MHz linear array transducers. In all cases, images of the biopsied nodules were obtained in transverse and longitudinal planes.
Pathological gold standard
Ultrasound images were obtained during routine scanning. FNA samples were obtained during standard clinical workflow and cytology was reviewed by pathology faculty at the institution. Determination of benignity or malignancy was made using FNA results, or when available, surgical specimens. For FNA, five categories were used: malignant, suspicious for malignancy, indeterminate, benign, and non-diagnostic. Only nodules that were malignant or benign were included in this study, unless a nodule underwent repeat FNA or surgical resection that subsequently provided confirmation of malignancy or benignity.
Image annotation
All images in the training set were interpreted by one of two ACR TI-RADS committee radiologists who were blinded to pathology results. The first reader (W.D.M.) had 22 years of experience and the second reader had 20 years of experience in thyroid imaging. Following the ACR TI-RADS lexicon, the readers assigned features for nodule composition, echogenicity, margins, and echogenic foci. For the echogenicity category, the readers classified 243 nodules as “moderate to markedly hypoechoic”, which was not compatible with the ACR TI-RADS lexicon. For these cases, a third reader (B.W.), a board-eligible radiology fellow with specialty practice in thyroid imaging and five years of experience, reviewed the echogenicity feature and modified it using the original assignment and additional imaging review. This reader also evaluated nodules for the shape feature. Eventually, all 1377 nodules were appropriately assigned all five ACR TI-RADS categories.
Annotations for the five ACR TI-RADS feature categories for the test nodules were performed by twelve radiologists in December 2016, before the publication of ACR TI-RADS, with the readers blinded to the pathology results. These interpretations were based on images obtained in transverse and longitudinal planes as well as video clips obtained in at least one plane displayed to the readers on a standard computer monitors using a website interface. Independent interpretations by three radiologists who were experts on the ACR TI-RADS committee, one of whom is a co-author (F.N.T), were combined into expert consensus using majority vote. These radiologists had between 26 and 34 years of post-training experience.
Among the remaining nine readers, one of them (W.D.M) was a member of the ACR TI-RADS committee with 22 years of experience and also interpreted the training cases. The other eight radiologists reported thyroid ultrasound in their clinical practice but had no knowledge of management recommendations in ACR TI-RADS. This group included two academic radiologists with sub-specialty training in ultrasound and 20 and 32 years of practice experience. The six remaining radiologists from this group were from private practices with fellowship training in neuroradiology, women’s imaging, and nuclear medicine, with experience ranging from 3 to 32 years.
Based on feature assessments for the five ACR TI-RADS categories from each reader, we first computed a total number of points per nodule and corresponding ACR TI-RADS risk levels. Then, according to ACR TI-RADS guidelines, we retrospectively decided whether a nodule would qualify for FNA and follow-up based on its size and ACR TI-RADS risk level.
Deep learning algorithm
Our proposed deep learning algorithm had three main stages: (1) nodule detection followed by (2) prediction of malignancy and (3) risk level stratification. Figure 2 presents these stages and how they are connected. A complete description of all the components of the deep learning algorithm are provided in Appendix 1.
Figure 2:
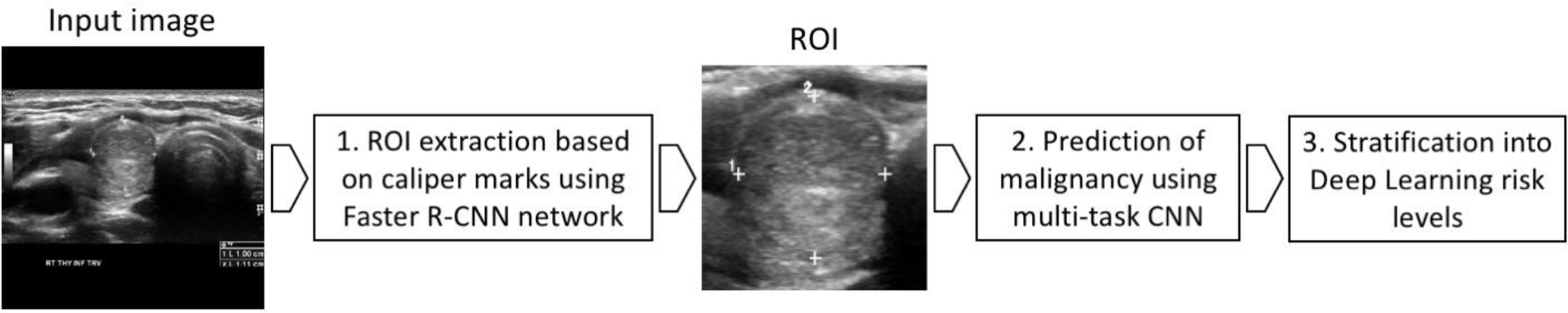
A flowchart presenting the three main processing stages of our deep learning algorithm. ROI = region of interest, CNN = convolutional neural network.
For nodule detection, we first obtained a bounding box of a nodule by enclosing calipers included in every image (used in clinical practice for nodule measurement). To detect the calipers, we trained a Faster R-CNN detection algorithm (13). After detecting the calipers within the ultrasound image, we extracted a square image with a fixed size margin of 32 pixels enclosing the corresponding nodule, resized the image to 160×160 pixels, and applied preprocessing (details in Appendix 1).
For classification, we trained from scratch a custom, multi-task deep convolutional neural network. The tasks used for training were presence or absence of malignancy and all of the ACR TI-RADS features across all five categories (composition, echogenicity, shape, margin, and echogenic foci). The architecture of our common representation extraction network is shown in Figure 3. Source code of the model is available at the following link: https://github.com/MaciejMazurowski/thyroid-us.
Figure 3:
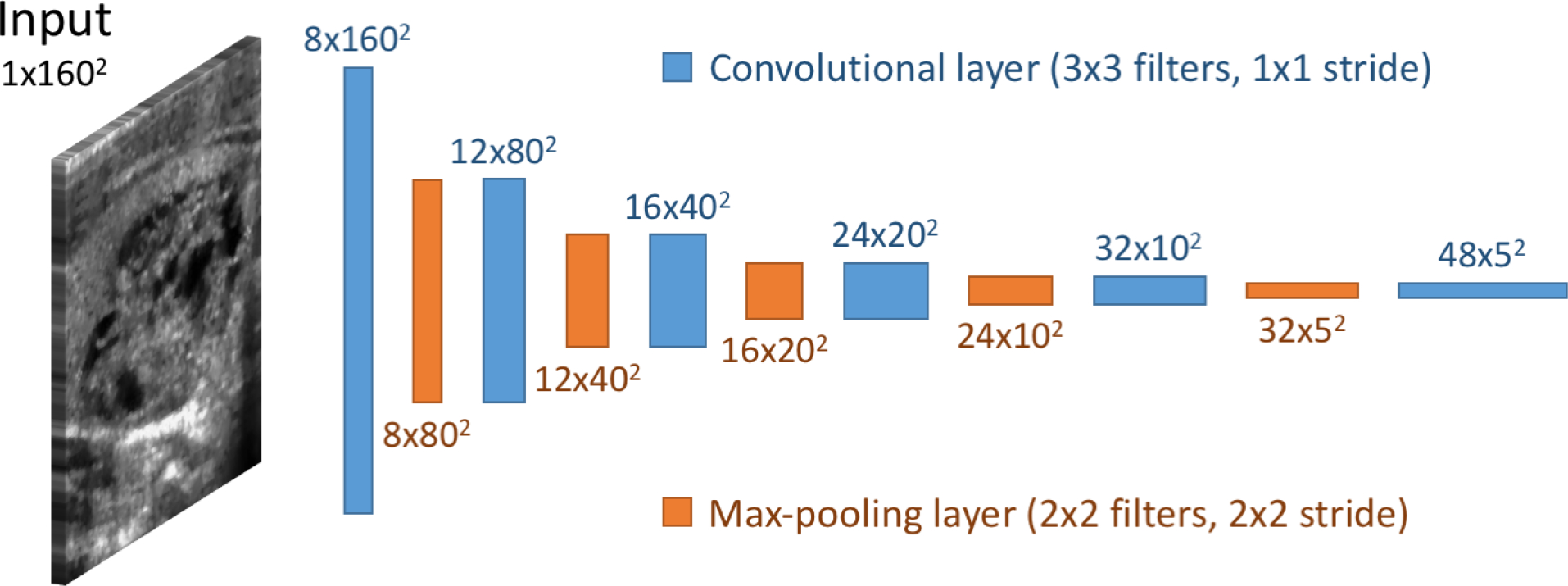
Convolutional neural network architecture of the network for shared representation extraction.
During inference, we stratified the probability of malignancy returned by the network into risk levels referred to as Deep Learning risk levels (DL2 – DL5), modeled after the ones defined in ACR TI-RADS (TR2 – TR5). Using the DL risk level and a nodule’s size resulted in a recommendation for FNA and follow-up. The size thresholds for FNA and follow-up recommendation were kept the same as in ACR TI-RADS. This step served the purpose of choosing appropriate point on ROC curve that takes into account nodule size and results in clinically relevant decisions.
Evaluation
We evaluated our deep learning algorithm and compared it to radiologists in two steps shown in Figure 4. First, we compared the performance of the algorithm to human readers in terms of discriminating benign and malignant nodules alone using the area under the receiver operating characteristic curve (AUC). This is the first and principal step of our algorithm as well as the ACR TI-RADS and it does not involve nodule size. The AUC for the deep learning algorithm was calculated using the likelihood of malignancy returned by model, and the AUC for radiologists used the total number of points computed with ACR TI-RADS. Then, for the second step, we evaluated the performance of the entire system in terms of sensitivity and specificity for recommendation of FNA and follow-up which, in addition to the first step described above, involves size-based thresholding. This two-step evaluation allows for isolating the predictive performance that is purely image-based from the final size-based recommendation step which aims to relate to the risk that malignant nodules of different sizes pose to patients.
Figure 4:
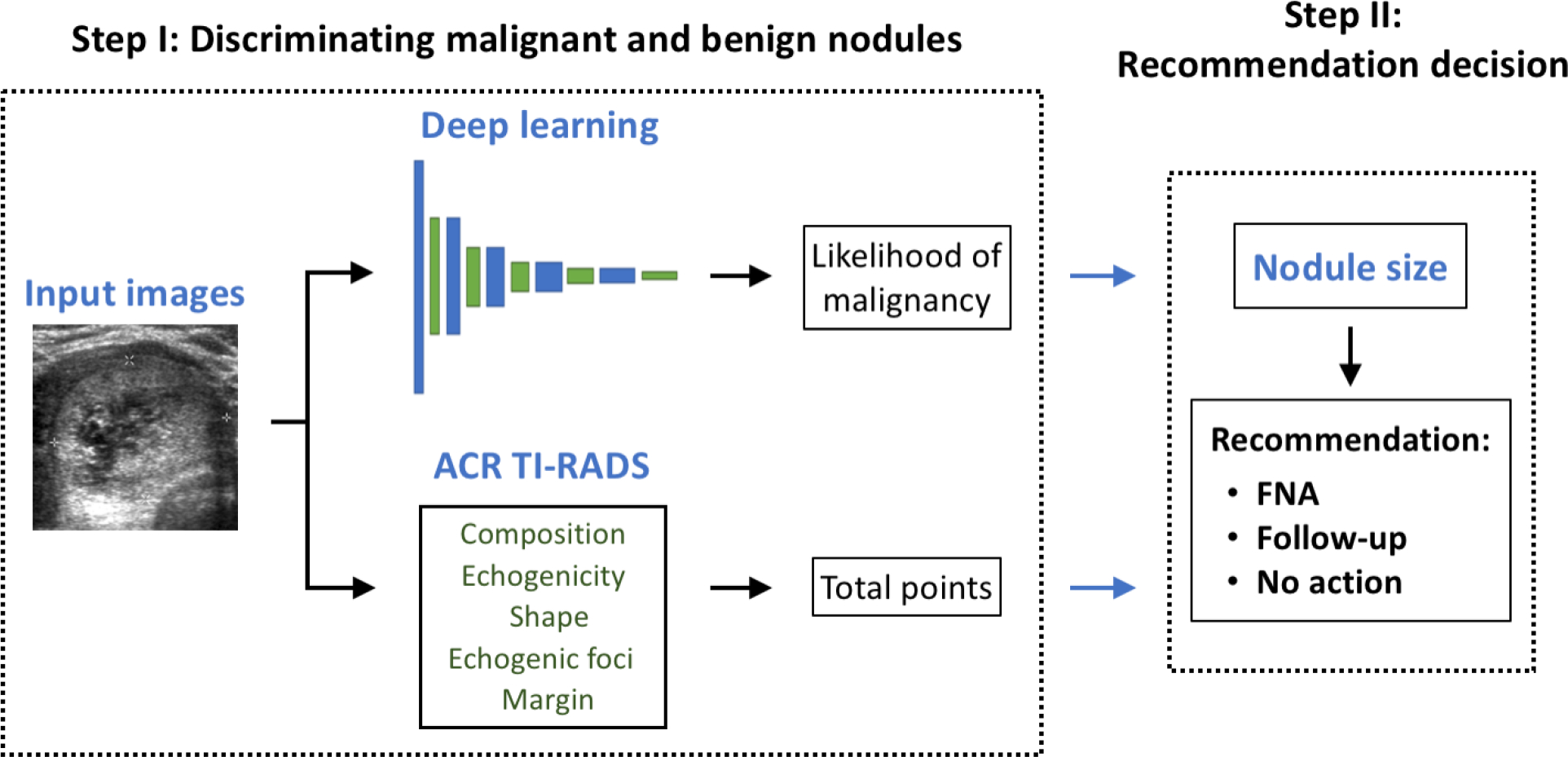
A diagram of the two-step decision making process for management of thyroid nodules. FNA = fine-needle aspiration.
We performed validation of the performance of the deep learning classifier in two ways: (1) using a 10-fold cross-validation with our training set by pooling predictions from all 10 non-overlapping folds and (2) using a held-out test set of 99 cases. On the training set, the deep learning algorithm was compared to a single radiologist in terms of AUC whereas on the test set we compared it to consensus of the three ACR TI-RADS committee members, and the nine other radiologists. Statistical tests for all comparisons were performed with bootstrapping.
RESULTS
Study population
The total number of malignant nodules was 142/1377 (10.3%) in all analyzed cases, with 127/1278 (9.9%) in the training set and 15/99 (15.2%) in the test set. The difference in the prevalence of malignant nodules between the training and test sets was not statistically significant (p>0.05). The mean maximum nodule size for all cases was 2.6 cm, with 2.6 cm in the training set and 2.7 cm in the test set (p>0.5). Statistics available for the entire population as well as for the training and test cases are found in Table 1.
Table 1:
Population statistics and the prevalence of malignant nodules class. SD = standard deviation.
| Findings | All cases (n=1377) | Training cases (n=1278) | Test cases (n=99) |
|---|---|---|---|
| Age in years (mean, SD, median) | 53.2, 14.0, 53 | 53.2, 13.9, 53 | 52.3, 14.0, 53 |
| Nodule size in cm (mean, SD, median) | 2.6, 1.5, 2.3 | 2.6, 1.5, 2.2 | 2.7, 1.3, 2.4 |
| Malignant nodules (n, %) | 142 (10.3%) | 127 (9.9%) | 15 (15.2%) |
Comparison of Deep Learning and Radiologists
The deep learning algorithm performed as follows on the task of discriminating benign and malignant nodules. On the training set of 1278 nodules, as evaluated using 10-fold cross-validation, the deep learning algorithm achieved an AUC of 0.78 (95% confidence interval [CI] 0.74–0.82) as compared to 0.80 (95% CI: 0.76–0.84) (p>0.4) for a single ACR TI-RADS committee radiologist using ACR TI-RADS (Figure 5a).
Figure 5a:
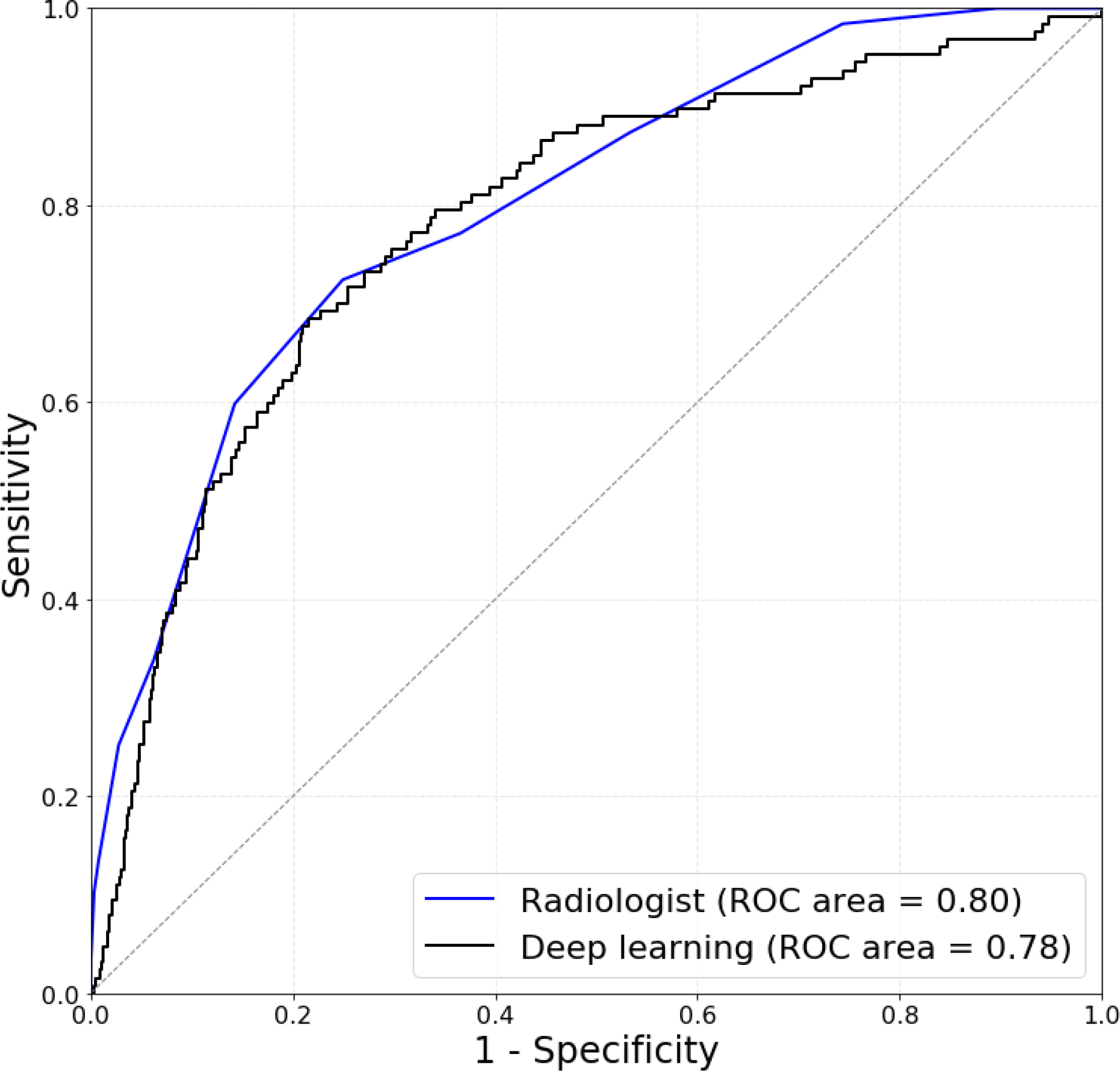
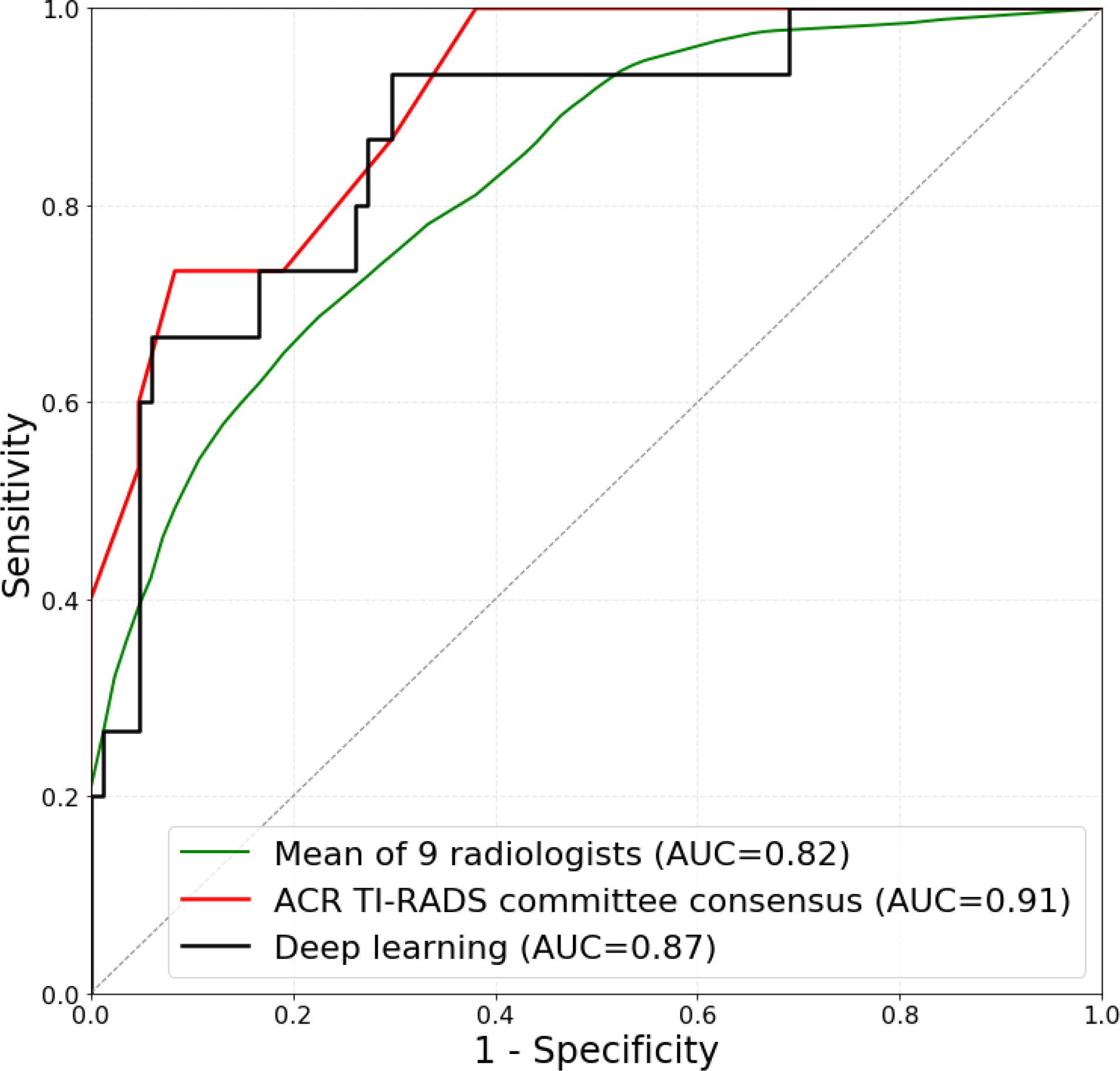
Receiver operating characteristic (ROC) curves comparing (a) deep learning evaluated using 10-fold cross-validation on 1278 training cases to a single radiologist following ACR TI-RADS and (b) deep learning evaluated on 99 test cases to expert consensus of three ACR TI-RADS committee members, and nine radiologists following ACR TI-RADS.
The performance of the algorithm and radiologists on the test set was as follows. For discriminating malignant and benign nodules, in terms of AUC, deep learning achieved 0.87 (95% CI: 0.76–0.95) which is comparable (p>0.4) to 0.91 (95% CI: 0.82–0.97) of expert consensus. The mean AUC of the nine radiologists was 0.83 (not significantly lower than for deep learning, p>0.3) with lowest being 0.76 (95% CI: 0.63–0.88) and highest 0.85 (95% CI: 0.76–0.94). Eight of the nine individual radiologists performed worse than deep learning, however, these differences were not statistically significant (p>0.05). The score of each reader is given in Table 2 and the mean ROC curve is shown in Figure 5b.
Table 2:
Comparison of the deep learning algorithm, consensus of three ACR TI-RADS committee expert readers, and nine radiologists on the test set of 99 nodules. FNA = fine-needle aspiration, ROC AUC = area under the receiver operating characteristic curve, SD = standard deviation.
| Reader | FNA |
Follow-up |
ROC AUC | Years of experience | ||
|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Sensitivity | Specificity | |||
|
| ||||||
| Deep learning | 13/15 (87%) | 44/84 (52%) | 14/15 (93%) | 32/84 (38%) | 0.87 | − |
| Expert consensus | 13/15 (87%) | 43/84 (51%) | 15/15 (100%) | 34/84 (40%) | 0.91 | 26–32 |
| Reader 1 | 14/15 (93%) | 40/84 (48%) | 15/15 (100%) | 28/84 (33%) | 0.91 | 20–25 |
| Reader 2 | 13/15 (87%) | 24/84 (29%) | 15/15 (100%) | 14/84 (17%) | 0.76 | 20 |
| Reader 3 | 12/15 (80%) | 40/84 (48%) | 15/15 (100%) | 27/84 (32%) | 0.85 | 13 |
| Reader 4 | 12/15 (80%) | 40/84 (48%) | 15/15 (100%) | 28/84 (33%) | 0.83 | 13 |
| Reader 5 | 11/15 (73%) | 49/84 (57%) | 14/15 (93%) | 34/84 (40%) | 0.78 | 3 |
| Reader 6 | 11/15 (73%) | 59/84 (70%) | 13/15 (87%) | 51/84 (61%) | 0.85 | 32 |
| Reader 7 | 12/15 (80%) | 42/84 (50%) | 15/15 (100%) | 33/84 (39%) | 0.81 | 4 |
| Reader 8 | 13/15 (87%) | 32/84 (38%) | 14/15 (93%) | 19/84 (23%) | 0.79 | 32 |
| Reader 9 | 14/15 (93%) | 37/84 (44%) | 15/15 (100%) | 26/84 (31%) | 0.83 | 20 |
| Readers 1–9 | 0.82 | |||||
| 83% (7.5%) | 48% (11.7%) | 97% (4.8%) | 34% (12.4%) | 17 (10) | ||
| mean (SD) | (0.05) | |||||
For FNA recommendations, after applying size thresholds, the sensitivity of the proposed deep learning algorithm was 13/15 (87%) (95% CI: 67–100%), the same as expert consensus with sensitivity of 13/15 (87%) (95% CI: 67–100%) and higher than five of the nine radiologists with sensitivity ranging from 11/15 (73%) to 14/15 (93%). The differences between sensitivity of deep learning and radiologists were not statistically significant (p>0.4). In terms of specificity, deep learning achieved 44/84 (52%) (95% CI: 41–63%) which was higher, although not significantly (p>0.5), than expert consensus (43/84 (51%) with 95% CI: 41–62%) and seven of the nine radiologists with specificity ranging from 24/84 (29%) to 59/84 (70%). The differences between specificity of deep learning and two out of these seven radiologists (Reader 2 and Reader 8) were statistically significant (p<0.05). The mean sensitivity and specificity for all nine radiologists was 83% and 48%, respectively, both lower than for the deep learning algorithm (p>0.4). Sensitivity and specificity for FNA recommendation by all readers is provided in Table 2. Out of the 42/99 nodules which were misclassified by deep learning, the nine radiologists misclassified 75% on average. In contrast, out of the nodules misclassified by radiologists (51.1/99 on average), deep learning misclassified 64%. This demonstrates a notable overlap in the misclassified cases and somewhat lower misclassification rate by the deep learning algorithm as compared to radiologists.
In recommending follow-up, deep learning performed similarly to all tested radiologists. Its sensitivity was 14/15 (93%) (95% CI: 78–100%). Expert consensus did not miss any malignant nodule for follow-up with the specificity of 34/84 (40%) (95% CI: 30–51%). Similar specificity was obtained by the deep learning algorithm (32/84 (38%) with 95% CI: 28–49%). For the remaining nine readers, the mean sensitivity was 97% whereas the mean specificity was relatively low (34%). In Table 2, we provide sensitivity and specificity for follow-up recommendation by all readers.
In addition, we split the positive (malignant) and negative (benign) test cases into two subsets, easy and difficult, based on human raters’ performance. 10 of 15 total positive cases were included in the easy set based on unanimous correct management decisions from all 10 readers (expert consensus and nine individual radiologists). 39 of 84 negative cases were also included in the easy set based on at least 6 out of 10 correct management decisions for FNA recommendation. These selections resulted in two subsets, one with 49 easy cases (10 positive and 39 negative) and the other with 50 difficult cases (5 positive and 45 negative). Figure 6 compares the performance of deep learning and radiologists on a subset of easy and difficult test cases. Deep learning achieved higher ROC AUC than radiologists on the difficult cases (0.92 versus 0.70) (p=0.021) and similar on the easy ones (0.89 versus 0.92) (p>0.5). When compared to expert consensus, on the difficult subset deep learning performed similarly (0.90 and 0.92, respectively) whereas on the easy subset, ROC AUC of deep learning (0.89) was slightly lower than for expert consensus (0.96). The differences between deep learning and expert consensus on both subsets were not statistically significant (p>0.1).
Figure 6:
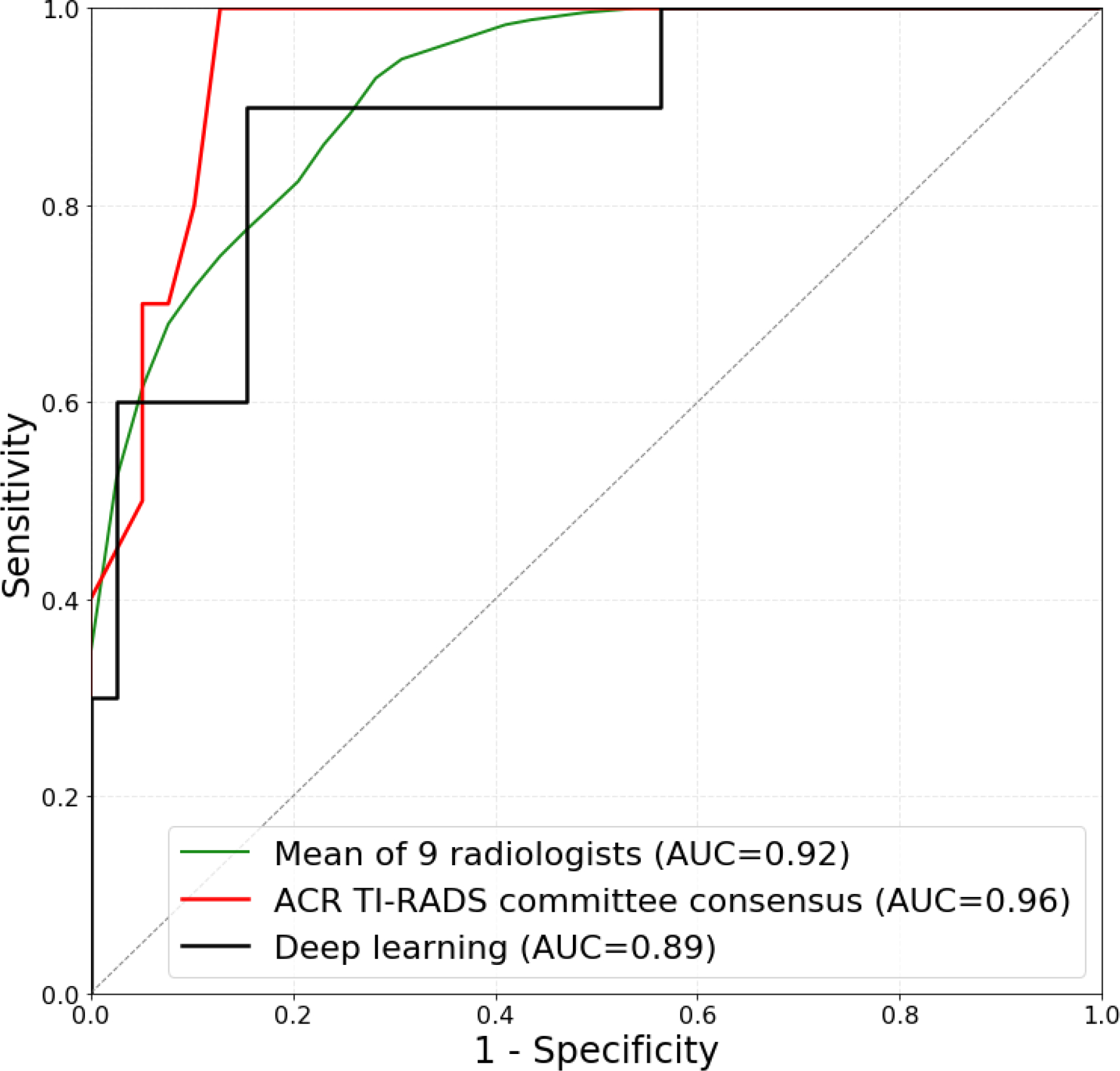
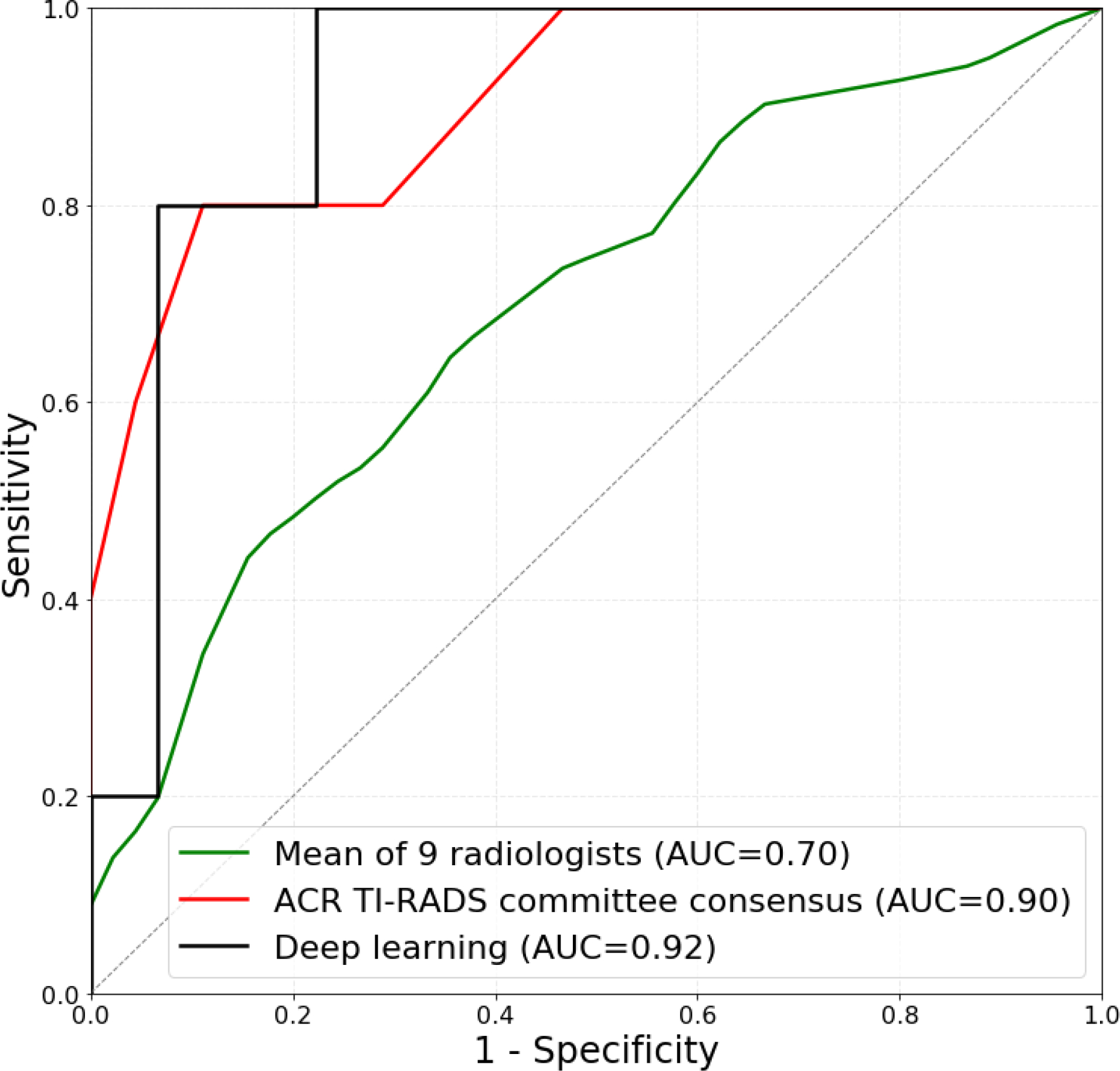
Receiver operating characteristic (ROC) curves comparing deep learning and radiologists using ACR TI-RADS on (a) 49 easy test cases and (b) 50 difficult test cases. Easy subset includes 10 malignant nodules based on unanimous correct management decisions from 9 readers and expert consensus and 39 benign cases based on at least 6 out of 10 correct management decisions for FNA recommendation. Difficult subset contains the remaining 5 malignant and 45 benign test cases.
DISCUSSION
In this study, we showed that a deep learning algorithm can match and, in some instances, exceed the performance of a radiologist on the task of recommending management for thyroid nodules seen on ultrasound images. Specifically, we tested the sensitivity and specificity of the algorithm for FNA and follow-up recommendation and showed that it performed similarly to radiologists following ACR TI-RADS guidelines.
The most valuable aspect of the deep learning algorithm is the ability to improve specificity of thyroid nodule biopsy recommendations. In a study that compared recommendations of eight radiologists for 100 nodules, Hoang et. al. found ACR TI-RADS to offer a meaningful reduction in the number of thyroid nodules recommended for biopsy and improved specificity (5). In our study, we show that deep learning maintains or could provide improvement in specificity compared to radiologists using ACR TI-RADS, which suggests that the proposed algorithm offers performance markedly higher than radiologists not using ACR TI-RADS. The latter is more likely in practice given application of the five features of ACR TI-RADS can be time consuming.
Our results add to the growing body of evidence demonstrating the potential power of deep learning when applied to thyroid ultrasound. Chi et. Al. showed that by utilizing imaging features extracted with a deep convolutional neural network they can achieve accuracy above 99% for the binary task of classifying thyroid nodules on ultrasound images to TI-RADS risk levels 1/2 vs. all others (14). Even though the performance seems to be outstanding, it refers to a greatly simplified task of predicting proxy labels. In contrast, our ground truth used for both training and testing relies on cytology and pathology results. In another study, Ma et al. used a large dataset of over 8000 thyroid nodules with malignant and benign status confirmed either by surgery or FNA result (15). The proposed deep learning algorithm that required manual nodule segmentation resulted in high sensitivity (82%) and specificity (84%), however, nodule sizes were not considered in evaluation. The malignancy rate was also high in this study and not reflective of a typical cohort of thyroid nodules undergoing thyroid ultrasound or biopsy. To the best of our knowledge, our study is the first that compares fully autonomous decisions made by a deep learning algorithm to radiologists.
A deep learning algorithm for prediction of malignancy could make a significant difference in a clinical practice. First, for a given image, our algorithm will always provide the same prediction. Therefore, it will eliminate a substantial inter-reader variability that has been observed for this task including in the TI-RADS system. Second, the algorithm could reduce the time required for interpretation of thyroid nodules which currently puts some strain on radiology departments. Finally, deep learning may perform better than some radiologists interpreting thyroid ultrasound in clinical practice, though a larger study is needed to confirm this.
Please note that the TI-RADS system consists of two steps. The first step, based on specific features of the nodules, estimates the likelihood that the lesion is malignant. The second step triages nodules for biopsy or follow-up based on the likelihood estimated in the first step as well as nodule size. Our deep learning system replaces only the first step and utilizes the same size-based triaging in the second stage. While this design decision was important to allow for a fair comparison of our system with TI-RADS in the proper clinical setting, it limits the system to some extent to the decision-making framework of TI-RADS. Future improvement that takes into account the interactions between tumor size and more detailed features of the nodules could provide additional gains in performance in terms of sensitivity and specificity.
Our study had some limitations. Specifically, our final test set of 99 nodules (with 15 positive and 84 negative cases) did not allow for a high confidence score and resulted in wide confidence intervals. This limitation was alleviated by a cross-validation experiment on the much larger training set (with 127 positive and 1151 negative cases), which showed results consistent with those on the test set (in terms of the comparable performance of our algorithm with the radiologist with the highest performance). Another limitation was that we noticed some differences in performance between the test set and the training set. This was not an indication of a high-bias model (underfitting) since it was the case for both deep learning and the radiologist. We believe that the main reason for this slight difference is that the nodules from the trainings set were on average more difficult which was corroborated by additional exploration of the data including evaluation of the discriminative power of features. Please note that while the overall performance of all predictors (deep learning and radiologists) differed between the two sets, the relative trends between radiologists and our algorithm remained. Regarding the study population, all nodules used in our study underwent FNA because of suspicious or indeterminate ultrasound findings and not based on ACR TI-RADS guidelines. This factor could account for relatively low specificity for all the readers and deep learning. In addition, no large-scale test set from external institution was available for comparison and to assess for generalization to broader population of patients and nodules.
In summary, in this paper we showed that deep learning algorithms are highly promising in the decision making process for thyroid nodules. Further studies are needed to further validate our findings.
Key Points.
We proposed a fully automatic deep learning system for providing biopsy recommendations for thyroid nodules and compare it to radiologists following ACR TI-RADS.
Our deep learning system achieved 52% specificity at 87% sensitivity in recommending biopsy for thyroid nodules compared to 51% specificity at 87% sensitivity by a consensus of three ACR TI-RADS committee experts.
ACKNOWLEDGEMENTS:
We thank the 12 radiologists who interpreted the test set of thyroid nodules as part of previously published work.
Appendix 1: Deep learning algorithm
Detection
For nodule detection, we first obtained a bounding box of a nodule by enclosing calipers included in every image (used in clinical practice for nodule measurement). To detect the calipers, we trained a Faster R-CNN detection algorithm (13) with ResNet-101 backbone network (16) initialized with weights pre-trained on the MS COCO detection dataset (17). It was fine-tuned using a training set of 2556 images with 7858 annotated calipers. The precision of our caliper-detection network was 98.82%. After detecting the calipers within the ultrasound image, we extracted a square image with a fixed size margin of 32 pixels enclosing the corresponding nodule. All images extracted this way contained the nodule of interest. Then, we resized the image to 160×160 pixels using bicubic interpolation and applied contrast normalization by removing the top and bottom 1% of pixel values. Finally, we performed median filter and non-local means algorithm (18), used for speckle noise reduction in US images (19), to improve classification results.
Classification
For classification, we trained from scratch a custom, multi-task deep convolutional neural network. The main idea of this method was to solve multiple tasks at once using a single model that was trained jointly for all of them. This was achieved by sharing a large portion of the network’s parameters between tasks to obtain a common representation. In multi-task learning, each task serves as a regularizer for the others, and all of them perform better when trained together rather than separately (20). The architecture of our common representation extraction network is shown in Figure 3. It was comprised of six blocks with 3×3 convolutional filters, followed by ReLU activation function and max pooling layer with 2×2 kernels. Instead of max pooling, the last block had a 50% dropout layer for additional regularization (21). Then, each task attached its own fully connected layer and softmax activation to obtain a class probabilities vector. The tasks used for training were presence or absence of malignancy and all of the ACR TI-RADS features across all five categories (composition, echogenicity, shape, margin, and echogenic foci). All tasks were either binary (malignancy and shape) or multiclass classification problems.
The network was trained for 250 epochs using RMSprop stochastic optimizer with batch size of 128. Optimization function for all tasks was a focal loss, suitable for imbalanced datasets (22,23). The overall loss was computed as a sum of six losses from all tasks. During the training phase, we used data augmentation by applying random rotation and sheer by 15 degrees, translation by 32 pixels, and scale by 10%. All hyperparameters were tuned on the training set of 1278 cases using a preliminary validation split comprising 10% (n=128) of randomly selected cases from this set.
Risk group stratification
During inference, we determined the probability of malignancy for each nodule by averaging the model prediction from both, transverse and longitudinal, images. Next, we stratified the probability of malignancy into risk levels referred to as Deep Learning risk levels (DL2 – DL5). These levels were modeled after the ones defined in ACR TI-RADS (TR2 – TR5), with the algorithm’s predictions for the training cases getting split into the same percentiles as their ACR TI-RADS risk levels. The thresholds for DL risk levels 3, 4, and 5 were 21.0%, 25.3%, and 34.7%, respectively. Using the DL risk level and a nodule’s size resulted in a recommendation for FNA and follow-up. The size thresholds for FNA and follow-up recommendation were kept the same as in ACR TI-RADS. This step served the purpose of choosing appropriate point on ROC curve that takes into account nodule size and results in clinically relevant decisions.
REFERENCES:
- 1.Haugen BR, Alexander EK, Bible KC, et al. 2015 American Thyroid Association Management Guidelines for Adult Patients with Thyroid Nodules and Differentiated Thyroid Cancer: The American Thyroid Association Guidelines Task Force on Thyroid Nodules and Differentiated Thyroid Cancer. Thyroid. 2016; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoang JK, Middleton WD, Farjat AE, et al. Interobserver Variability of Sonographic Features Used in the American College of Radiology Thyroid Imaging Reporting and Data System. Am J Roentgenol. 2018;1–6. [DOI] [PubMed] [Google Scholar]
- 3.Vaccarella S, Franceschi S, Bray F, Wild CP, Plummer M, Dal Maso L. Worldwide Thyroid-Cancer Epidemic? The Increasing Impact of Overdiagnosis. N Engl J Med. 2016;375(7):614–617. [DOI] [PubMed] [Google Scholar]
- 4.Tessler FN, Middleton WD, Grant EG, et al. ACR thyroid imaging, reporting and data system (TI-RADS): white paper of the ACR TI-RADS committee. J Am Coll Radiol. Elsevier; 2017;14(5):587–595. [DOI] [PubMed] [Google Scholar]
- 5.Hoang JK, Middleton WD, Farjat AE, et al. Reduction in thyroid nodule biopsies and improved accuracy with American college of radiology thyroid imaging reporting and data system. Radiology. 2018;287(1). [DOI] [PubMed] [Google Scholar]
- 6.Griffin AS, Mitsky J, Rawal U, Bronner AJ, Tessler FN, Hoang JK. Improved Quality of Thyroid Ultrasound Reports After Implementation of the ACR Thyroid Imaging Reporting and Data System Nodule Lexicon and Risk Stratification System. J Am Coll Radiol. Elsevier; 2018;15(5):743–748. [DOI] [PubMed] [Google Scholar]
- 7.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. Nature Publishing Group; 2015;521(7553):436. [DOI] [PubMed] [Google Scholar]
- 8.Esteva A, Kuprel B, Novoa R, Ko J, Nature SS-, 2017 U. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. Jama. American Medical Association; 2016;316(22):2402–2410. [DOI] [PubMed] [Google Scholar]
- 10.Erickson BJ, Korfiatis P, Akkus Z, Kline TL. Machine Learning for Medical Imaging. RadioGraphics. Radiological Society of North America; 2017;37(2):505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee H, Tajmir S, Lee J, et al. Fully automated deep learning system for bone age assessment. J Digit Imaging. Springer; 2017;30(4):427–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazurowski MA, Buda M, Saha A, Bashir MR. Deep learning in radiology: An overview of the concepts and a survey of the state of the art with focus on MRI. J Magn Reson Imaging. Wiley Online Library; 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren S, He K, Girshick R, Sun J. Faster R-CNN: Towards real-time object detection with region proposal networks. Adv Neural Inf Process Syst. 2015. p. 91–99. [DOI] [PubMed] [Google Scholar]
- 14.Chi J, Walia E, Babyn P, Wang J, Groot G, Eramian M. Thyroid Nodule Classification in Ultrasound Images by Fine-Tuning Deep Convolutional Neural Network. J Digit Imaging. 2017;30(4):477–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma J, Wu F, Zhu J, Xu D, Kong D. A pre-trained convolutional neural network based method for thyroid nodule diagnosis. Ultrasonics. Elsevier; 2017;73:221–230. [DOI] [PubMed] [Google Scholar]
- 16.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. Proc IEEE Conf Comput Vis Pattern Recognit. 2016. p. 770–778. [Google Scholar]
- 17.Lin T-Y, Maire M, Belongie S, et al. Microsoft COCO: Common objects in context. Eur Conf Comput Vis. 2014. p. 740–755. [Google Scholar]
- 18.Buades A, Coll B, Morel J-M. A non-local algorithm for image denoising. Comput Vis Pattern Recognition, 2005 CVPR 2005 IEEE Comput Soc Conf. 2005. p. 60–65. [Google Scholar]
- 19.Coupé P, Hellier P, Kervrann C, Barillot C. Nonlocal means-based speckle filtering for ultrasound images. IEEE Trans image Process. IEEE; 2009;18(10):2221–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caruana R Multitask learning. Learn to Learn. Springer; 1998. p. 95–133. [Google Scholar]
- 21.Nitish Srivastava, Hinton Geoffrey E, Krizhevsky Alex, Sutskever Ilya SR. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014;15(1):1929--1958. [Google Scholar]
- 22.Lin T-Y, Goyal P, Girshick R, He K, Dollár P. Focal loss for dense object detection. arXiv Prepr arXiv170802002. 2017; [DOI] [PubMed] [Google Scholar]
- 23.Buda M, Maki A, Mazurowski MA. A systematic study of the class imbalance problem in convolutional neural networks. Neural Networks. Elsevier; 2018;106:249–259. [DOI] [PubMed] [Google Scholar]


