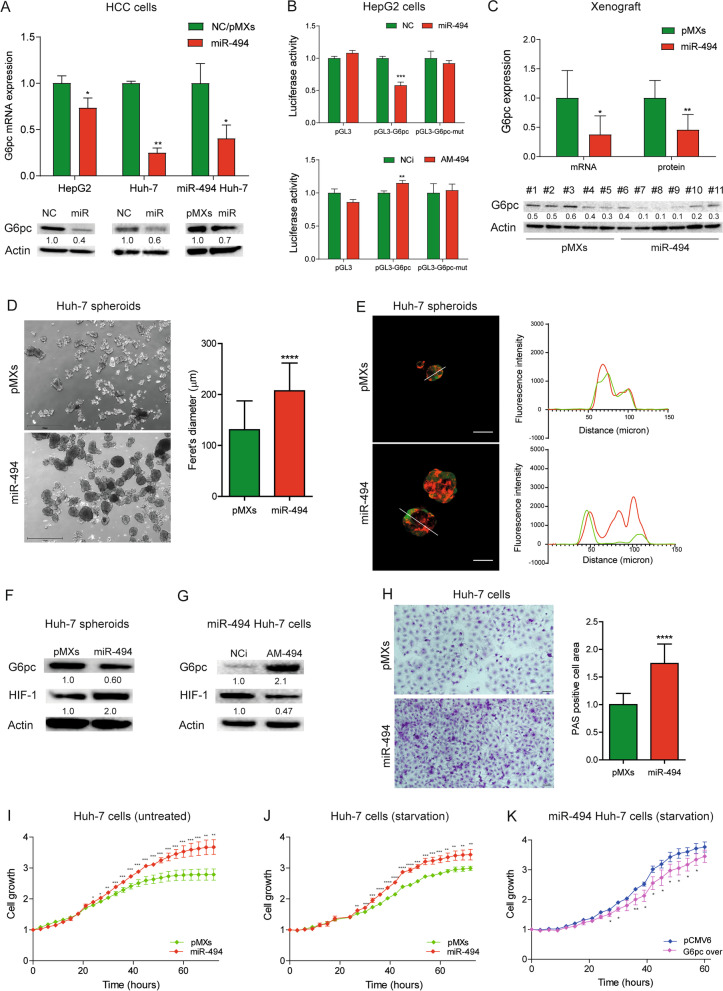
Fig. 2.
G6pc is a target of miR-494 in HCC. A Real Time PCR and WB analyses of G6pc expression following miR-494 overexpression in transfected HCC cells and infected Huh-7 cells (miR-494 Huh-7). NC: negative control precursor miRNA. PMXs: control vector. Y-axis reports 2−ΔΔ.Ct values corresponding to G6pc mRNA levels normalized to controls (NC or pMXs). Mean ± SD values are displayed. Beta-actin was used as housekeeping gene for Real Time and WB experiments. Real Time PCR analysis was performed in two independent experiments in triplicate; WB analysis was performed in two independent experiments. B Dual-luciferase activity assay of wild type and mutant (mut) G6pc-3UTR vectors (pGL3-G6pc) co-transfected with miR-494 or anti-miR-494 (AM-494) in HepG2 cells. NC: negative control precursor miRNA. NCi: negative inhibitor control miRNA. Y-axes report the firefly/renilla ratio normalized to controls (NC or NCi). Mean ± SD values are displayed. Analysis was performed in two independent experiments in triplicate. C Real Time PCR and WB analyses of G6pc expression in tumor masses (N = 16) of xenograft mice obtained following subcutaneous injection of miR-494-overexpressing and control (pMXs) Huh-7 cells. Y-axis reports G6pc mRNA and protein levels normalized to control. Mean ± SD values are displayed. Beta-actin was used as housekeeping gene for Real Time and WB experiments. Real Time PCR was run in triplicate. D Representative images (4X magnification) of miR-494-overexpressing and control (pMXs) Huh-7 spheroids at 24 h. Data were obtained by measuring feret’s diameter (µm) of thirty randomly selected spheroids in two independent experiments. Mean ± SD values are displayed. Scale bars, 750 μm. E Representative confocal images of control (pMXs) and miR-494-overexpressing Huh-7 spheroids expressing GFP (green signal) and stained with the live-cell oxygen sensor BTP (red signal), which fluorescence emission is quenched by molecular oxygen. Traces represent red and green fluorescence intensity along the linear regions of interest traced in the merged images to visualize signal distribution. At least ten spheroids for condition were analyzed in two independent experiments. Scale bars, 25 μm. F WB analysis of G6pc and HIF-1 in Huh-7 spheroids and (G) miR-494-overexpressing Huh-7 cells following AM-494 transfection. NCi: negative inhibitor miRNA control. PMXs: control vector. Beta-actin was used as housekeeping gene. WB analysis was performed in two independent experiments. H Representative images (20X magnification) of PAS staining in control (pMXs) and miR-494-overexpressing Huh-7 cells. Y-axis reports the percent of PAS positive cell area normalized to control. Mean ± SD values are displayed. Five randomly selected fields were analyzed from three independent experiments. Scale bars, 20 μm. I, J Growth curves of control (pMXs) and miR-494-overexpressing Huh-7 cells in standard and starved culture conditions. Growth curves were normalized to T0. Mean ± SD values are reported. Two independent experiments were performed in quadruplicate. K Growth curves of miR-494-overexpressing Huh-7 cells transfected with G6pc overexpressing (G6pc over) or control (pCMV6) vector and grown in starved (medium without FBS) culture conditions. Growth curves were normalized to T0. Mean ± SD values are reported. Two independent experiments were performed in quadruplicate. Statistical significance was determined by two-tailed unpaired Student's t-test. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001