Abstract
While bone marrow-derived mesenchymal stem cells (BM-MSCs) are widely recognized as promising therapeutic agents, the age-related impacts on cellular function remain largely uncharacterized. In this study, we found that BM-MSCs from young donors healed wounds in a xenograft model faster compared to their aged counterparts (p < 0.001). Given this significant healing advantage, we then utilized single-cell transcriptomic analysis to provide potential molecular insights into these observations. We found that the young cells contained a higher proportion of cells characterized by a higher expression of genes involved in tissue regeneration. In addition, we identified a unique, quiescent subpopulation that was exclusively present in young donor cells. Together, these findings may explain a novel mechanism for the enhanced healing capacity of young stem cells and may have implications for autologous cell therapy in the extremes of age.
Keywords: Aging, mesenchymal stem cells, regeneration, subpopulation, therapy
INTRODUCTION
Age-related impairment of stem cell function has been widely implicated in the functional deterioration of numerous organs and tissues such as the neural [1, 2], cardiac [3–5], and hematopoietic [6–8] systems (reviewed in [9, 10]). Reversing the aging effects on stem cell populations has shown promise and increased therapeutic potential [11–14]. In a fashion similar to that of organ-specific stem cells in maintaining organ homeostasis [15], mesenchymal stem cells (MSCs) exhibit regenerative properties and are actively recruited to sites of injury [16]. MSCs have increasingly been utilized for cell-based therapies in a wide variety of diseases where they have shown to be enhancers of tissue repair [17–21]. It has been further demonstrated that MSCs exert their pro-regenerative properties predominantly via paracrine cytokine excretion, microvesicles and subsequent stimulation of local cells responsible for tissue healing [22–24].
Studies investigating age-related effects on bone marrow-derived MSCs (BM-MSCs) have yielded inconsistent results [25, 26]. Some groups have reported no age-related differences in functions including differentiation potential and proliferation, while others have observed reduced proliferative [26, 27], anti-inflammatory [28] and multipotent [25] potential. To date, we still lack a clear understanding of the impact of aging on MSC populations.
Advances in microfluidic and molecular biology have allowed for the evaluation of cellular heterogeneity [29] within seemingly homogenous stem cell populations [30–32]. We have developed and validated a method that combines high throughput single cell gene expression analysis with complex mathematical modeling to identify differences in cell subpopulation dynamics [33], which we have previously used to characterize heterogeneous stem cell subpopulations [30, 34]. Applying this approach not only enables the detection of subpopulations that share a common gene-expression profile, but also provides insights into the regulatory and signaling landscape at single cell resolution by highlighting patterns of gene expression.
METHODS
Isolation, In Vitro Culture and Characterization of Human BM-MSCs
Human BM-MSCs were isolated, cultured and characterized as described previously (Siegel et al. 2013) from young (n = 3; μ = 24 years, σ = 8.3 years) and aged (n = 5; μ = 73.0 years, σ = 3.7 years) donors. After written informed consent was obtained and the local Institutional Review Board approved the study, BM was harvested from these patients during orthopedic procedures. BM mononuclear cells were isolated by density gradient technique (Lymphoflot ®, Biotest, Dreieich, Germany) and seeded with MSC culture medium into tissue culture flasks (Nunc, Roskilde, Denmark) at 1×105 cells/cm2. MSC culture medium was composed of α-MEM (Lonza, Portsmouth, NH, USA), 1% penicillin-streptomycin (Lonza) and 10 % pooled human serum (ZKT, Tübingen, Germany). Non-adherent cells were removed after 24 hours, after which the remaining adherent cells were cultured at 37°C in humidified atmosphere with 5% CO2.
Flow Cytometry
The following PE-labeled or unlabeled antibodies were used (secondary antibody: PE-conjugated goat anti-mouse Ig [BD Bioscience, San Jose, CA, USA]): anti-CD10, −CD14, −CD19, −CD29, −CD34, −CD43, −CD44, −CD45, −CD59, −CD71, −CD73, −CD90, −CD105, −CD106, −CD119, −CD130, −CD140a, −CD140b, −CD146, −CD166, −GD2 (BD Biosciences); -CD133, -CD271 (Miltenyi Biotec, Bergisch Gladbach, Germany); and –MSCA-1 (TNAP) (BioLegend, San Diego, CA, USA). PE-conjugated or non-labeled IgG1 and IgG2a antibodies (BD Bioscience) were used as isotype matched controls. Flow cytometry was performed with a FACScan instrument (BD, Franklin Lakes, NJ, USA) and BD CellQuest Pro™ software. Dead cells were identified by uptake of 7-Aminoactinomycin D. FlowJo-7.2.5 software (Tree Star, Ashland, OR, USA) was used for analysis.
In vitro Differentiation Assays
For adipogenic differentiation the hMSC Adipogenic BulletKit (PT-3004, Lonza) was used, and for osteogenic differentiation 10–8 M dexamethasone, 0.2 mM ascorbic acid and 10 mM β-glycerolphosphate (Sigma) was added to the media. For chondrogenic differentiation the hMSC Chondrogenic Differentiation BulletKit (PT-3003, Lonza) with TGF-β 3 (PT-4124, Lonza) was applied. Differentiation was assessed with specific stainings (Oil Red O, adipogenic; Alizarin Red, osteogenic; Safranin O, chondrogenic).
In Vivo Excisional Wound Model
16-week-old C57BL/6 mice with leptin receptor deficiency were randomized into three treatment groups; young human bone marrow Mesenchymal Stem Cells (BM-MSCs), aged human BM-MSCs, and murine fibroblasts as a cell control (n=5 mice per group). Two full-thickness wounds both 6 millimeter (mm) in diameter were excised on each side of the dorsal midline, after which each wound was splinted with silicone splints that were sutured in place to prevent wound contracture. 2×105 cells were administered to each wound bed after wound creation. All wounds were covered with a sterile occlusive dressing (Tegaderm, 3M, St. Paul, MN, USA). Digital photographs were taken on day 0 and 1 and every other day thereafter until full wound closure. The wound areas were measured using Adobe Photoshop CS6 (Adobe Systems, San Jose, CA, USA).
Immunohistochemistry
Wounds from the excisional model were harvested upon closure (day 24) and immediately fixed in 4% paraformaldehyde overnight, dehydrated with sequential ethanol concentrations (30, 50, 70, and 95%), xylene, and paraffin washes, and embedded in paraffin for sectioning. To evaluate dermal integrity, wound sections were stained with Masson’s Trichrome and visualized using light microscopy. To assess vasculature in the healed wounds, sections were stained for CD31 (1° - 1:200 Rb α CD31, Ab28364, Abcam; 2° - 1:200 AF488 Gt α Rb, Life Technologies, Carlsbad, CA, USA) and VEGF-A (1° - 1:200 Rb α VEGFA, Ab46154, Abcam; 2° - 1:200 AF594 Gt α Rb, Life Technologies). To assess superoxide protein levels in the healed wounds, sections were stained for SOD2 (1° - 1:200 Rb α SOD2, Ab13533, Abcam; 2° - 1:200 AF594 Gt α Rb, Life Technologies). To assess the presence of transplanted cells in the healed wounds, sections were stained for human nuclei (α -Nuclei, AF488 conjugate, MAB1281A4, EMD Millipore, Burlington, MA). For IHC analysis, nuclei were stained with DAPI, and Adobe Photoshop CS6 (Adobe Systems) was used to superimpose the images taken with the same settings. Stained pixels per high magnification field were quantified using ImageJ.
Microfluidic Single-Cell Gene Expression Analysis
Young and aged primary BM-MSCs with the surface marker profile CD45−/CD34−/CD90+, CD73+, CD105+ (see Supplementary figure 1A–B for a comprehensive profile) were sorted as single cells using a Becton Dickinson FACSAria flow cytometer into 6 μL of lysis buffer. Reverse transcription and low cycle pre-amplification were performed using Cells Direct (Invitrogen, Carlsbad, CA, USA) with Taqman assay primer sets (Applied Biosystems, Foster City, CA, USA) as per the manufacturer’s specifications. cDNA was loaded onto 96.96 Dynamic Arrays (Fluidigm, San Francisco, CA, USA) for qPCR amplification using Universal PCR Master Mix (Applied Biosystems) with a uniquely compiled Taqman assay primer set as previously described (32).
Analyses of trophic factors in vitro production and secretion
For in vitro analyses of trophic factors proteins MSCs (n=5 donors per group, P3-P5) were seeded at 20,000 cells/cm2 in α-MEM supplemented with 16.5% fetal calf serum and 1% penicillin-streptomycin. After 24h supernatants and cells were harvested, and after cell lysis (PBS+1%TritonX100), supernatants and cell lysates were stored at −80°C until analysis with enzyme-linked immunosorbent assays (ELISAs). The following ELISA kits were used according to the manufacturerś instructions: human vascular endothelial growth factor (VEGF)-A, human transforming growth factor (TGF)-ß, (both R&D Systems, Minneapolis, MN, USA), human superoxide dismutase (SOD) 1 (supernatants: Thermo Fisher, Waltham, MA, USA; lysates: RayBiotech, Norcross, GA, USA), and total protein BCA assay (Thermo Fisher). Factor concentrations were normalized to respective protein concentrations.
Data Analysis
Analysis of the single-transcripts was performed employing a two-sample Kolmogorov-Smirnov (K-S) test to compare empirical distributions, employing a strict cutoff of p < 0.01. Transcriptionally defined subpopulations were determined using a k-means based clustering algorithm. Each cell was assigned an individual cluster identity based on its gene expression profiles and subsequently partitioned. The partitioned clusters were then further stratified using hierarchical agglomerative clustering in order to facilitate data visualization and data analysis within and across these clusters using the Euclidean distance as the calculation metric with complete linkage. The heatmap, dendrogram, and t-SNE images were generated using MATLAB version R2015b (Mathworks, Natick, MA, USA). Canonical pathway calculations and network analyses were performed using Ingenuity Pathway Analysis (IPA, Ingenuity Systems, Redwood City, CA, USA) based on genes differentially expressed among cell subpopulations. Immunohistochemistry data (5 wounds per group analyzed) and ELISA data were statistically analyzed with unpaired Studentś t-test.
RESULTS AND DISCUSSION
To evaluate differences in the regenerative potential of human BM-MSCs from young and aged donors we first confirmed the identity and evaluated the health of MSC preparations from 5 aged (mean age: 73 years ± 3.7 years SD) and 3 young (mean age: 24 years ± 8.3 years SD) donors. We characterized the surface marker expression of using flow cytometry and found that all cells were CD73+/CD90+/CD105+/CD14−/CD19−/CD34−/CD45−, consistent with the surface marker profile of MSCs as defined by the International Society for Cellular Therapy (34) (Supplementary Figure 1). MSC preparations from young and aged donors demonstrated no difference in metabolic activity or proliferation rates and were capable of trilineage differentiation (Supplementary Figure 2).
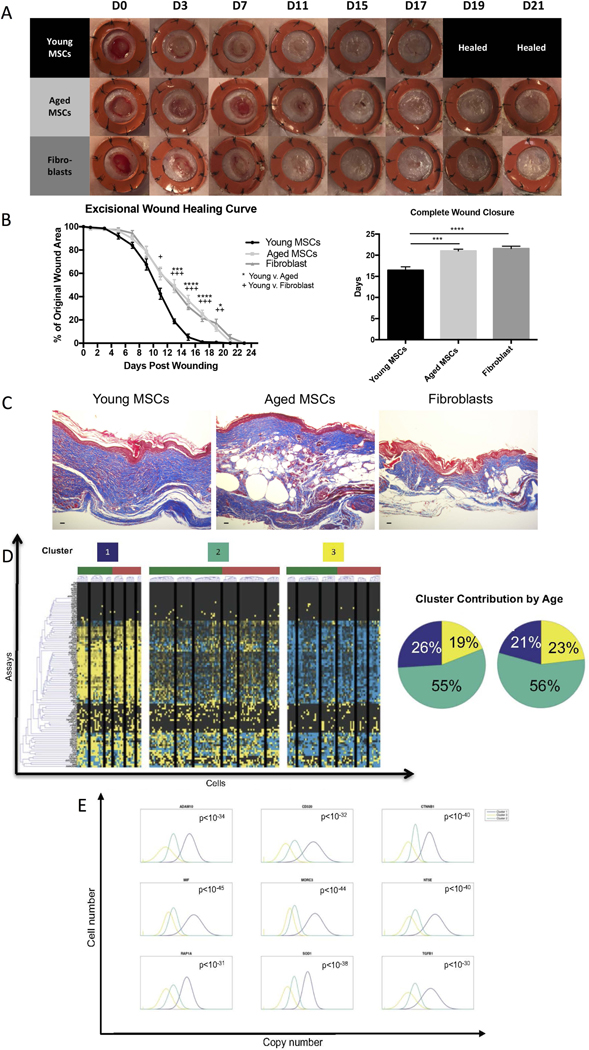
We next compared the wound healing ability of MSC preparations pooled from the young and aged donors. Our findings confirm a significant age-associated delay in the healing kinetics of BM-MSCs in a standard wound healing model (Figure 1A). Young BM-MSCs were able to establish complete skin formation over the wound almost five days earlier than aged BM-MSCs and fibroblasts (young BM-MSC: 16.43 ± 0.84 days, n = 5; aged BM-MSC: 21.00 ± 0.44 days, n = 5, p < 0.G001; fibroblast: 21.57 ± 0.57 days, n = 5; p ≤ 0.0001) (Figure 1B). The harvested wounds from the young BM-MSCs exhibited a more robust dermal layer and increased neovascularization (Figure 1C, Supplementary Figure 3A). At the time of wound closure no human cells were present anymore (not shown) but in both groups VEGF-A and SOD2 could be detected in the tissue (Supplementary Figure 3B). Both young and aged BM-MSCs produced and secreted VEGF-A, TGF-β and SOD1 in vitro (Supplementary Figure 3C).
Figure 1: Aged BM-MSCs exhibit a less regenerative phenotype and young BM-MSCs contain a higher proportion of cells belonging to a subpopulation correlated with a more regenerative molecular phenotype.
A-B) Murine diabetic wounds treated with a single, local injection of young BM-MSCs heal five days earlier than aged BM-MSCs and fibroblasts (young BM-MSCs: 16.43 ± 0.84, n = 5; aged BM-MSCs: 21.00 ± 0.44, n = 5, p < 0.001; fibroblasts: 21.57 ± 0.57, n = 5; p ≤ 0.0001). Error bars indicate mean ± SEM. C) Murine diabetic wounds treated with young BM-MSCs, aged BM-MSCs or fibroblasts harvested on the day of wound closure. Scale bar = 25 μm. D) Transcriptional profiles of young and aged BM-MSCs were evaluated using a microfluidic-based single-cell gene expression analysis platform to interrogate individual cells for 96 genes associated with migration, proliferation, stemness, and tissue regeneration. Black lines within heatmap divide K-means clustering of gene expression data reveals three transcriptionally distinct BM-MSC clusters, where Cluster 1 is characterized by significant upregulation of genes involved in pro-regenerative processes. Cluster pie charts representing the fraction of cells comprising each cluster show a decrease in the contribution of aged (red) BM-MSCs to Cluster 1 compared to young (green) BM-MSCs, suggesting an age-related shift in subpopulation dynamics. E) Top 9 Cluster 1 defining genes determined via non-parametric two-sample Kolmogorov-Smirnov testing. 50 genes exhibited significantly different (p < 0.01) distributions of single-cell expression in Cluster 1 (purple) in relation to the other clusters. (For full list see Supplemental Table 2).
To identify putative BM-MSC subpopulations potentially responsible for these discrepancies, we employed the previously described microfluidic-based single-cell gene expression analysis platform (32) to evaluate the transcriptional profiles of single cells simultaneously for 96 genes associated with cell migration, proliferation, stemness, and tissue regeneration (Figure 1D; Supplemental Table 1). Single cells can be reliably grouped together by their gene expression profiles when subjected to unsupervised clustering (35). In order to investigate the differences in expression profiles of young and aged BM-MSCs, we utilized an unsupervised k-means clustering algorithm to identify distinct cellular subsets (32) and detected three distinct BM-MSC subpopulation clusters (Figure 1D; Supplementary Figure 4). Cluster 1 was characterized by significant upregulation of genes responsible for cell recruitment, and free radical scavenging, such as beta-catenin (CTNNB1), macrophage inhibitory factor (MIF), and SOD (Figure 1E, full list: Supplemental Table 2). Interestingly, this high functionality cluster contained more young cells than aged cells, whereas Cluster 3, characterized by overall lower expression of genes in relation to the other clusters, was comprised of more aged cells (Figure 1D), suggesting an age-related shift in subpopulation dynamics. Not surprisingly, over 55% of cells from both the young and aged donors clustered together (Cluster 2), suggesting that age does not impact the majority of BM-MSCs.
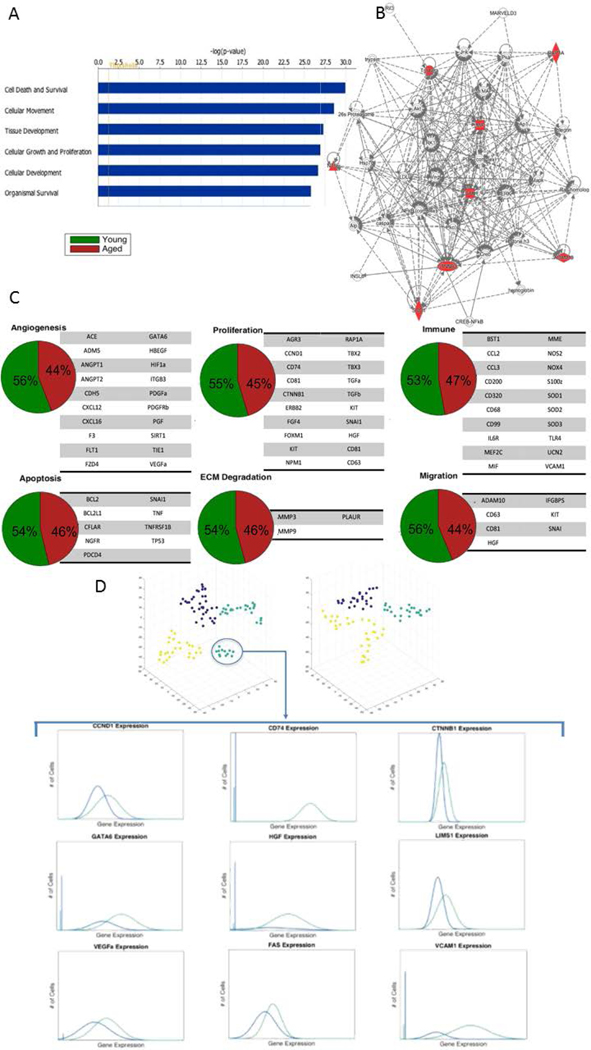
We subsequently investigated how age affects key cellular signaling networks. Utilizing MSC function-defining genes of the highly functional Cluster 1 as a source, we determined inferred canonical signaling pathways by interrogating Ingenuity Pathway Analysis (IPA, QIAGEN, Redwood City, CA, www.qiagen.com/ingenuity). Significantly upregulated pathways included cell survival, cellular movement, tissue development, immune cell trafficking, cellular development, as well as the degradation of superoxide radicals (Figure 2A, B; Supplementary Figures 4-6); supporting a highly regenerative profile of these cells.
Figure 2: Aged BM-MSCs are characterized by altered gene expressional subpopulation dynamics and a loss of a distinct quiescent cell population.
A) Top canonical signaling pathways upregulated in Cluster 1 inferred using Ingenuity Pathway Analysis (For full list see Supplementary Figure 5). B) Top scoring IPA-constructed transcriptome network based on Cluster 1 defining genes. Significant ‘seed’ genes are colored in red to distinguish them from the remaining ‘inferred’ entities in the network. C) Pie charts representing the fraction of young and aged MSCs expressing gene expression patterns related to pro-regenerative processes. Young BM-MScs exhibit only mildly increased gene expression signature in favor of the pro-regenerative phenotype. D) T-Distributed stochastic neighbor embedding (t-SNE)-based analysis (top) identifies a quiescent cell population (teal) within young BM-MSCs. Non-parametric two-sample Kolmogorov-Smirnov testing was used to identify genes defining this quiescent cell population and reveals that this subpopulation has overall diminished gene expression, particularly in genes responsible for proliferation and differentiation.
Having established that human BM-MSCs are a heterogeneous cell population and that profound differences in subpopulation ecology between young and aged donors exist, we next investigated how age-related BM-MSC subpopulations impact key processes of tissue repair. We applied analytical methods to compare gene expression patterns related angiogenesis, immunomodulation, migration, proliferation, apoptosis and extracellular matrix degradation (Figure 2C).
Interestingly, young BM-MSCs exhibited only a mildly increased gene expression signature in favor of the pro-regenerative phenotype. Skeptical of how this small gene expression advantage within the young BM-MSC population was able to elicit such a striking functional outcome in wound healing rate, we sought to identify other potential explanations for our in vivo observations. Hence, we applied a t-Distributed Stochastic Neighbor Embedding (t-SNE)-based analysis to the same raw data to explore the possibility of other patterns. T-SNE is a dimensionality reduction technique that provides an alternative means of data visualization (36). Upon application of t-SNE, we discovered a subpopulation of BM-MSCs with overall diminished gene expression, particularly with respect to genes responsible for proliferation and differentiation. Such genes included GATA6, LIM, LIMS1, and cyclinD1 (CCND1), a gene responsible for the progression of cells into the G1/S phase of the cell cycle. This minimal basal activity is indicative of quiescent cells and, most notably, was only present in the young BM-MSC population (Figure 2D). While quiescence is not definitive of stem cells, it is well established that loss of quiescence often results in stem cell depletion by favoring an imbalance of progenitor cell populations. As we did not observe obvious differences of selected trophic factors on protein level between the complete, i.e. heterogeneous, BM-MSC preparations from young and aged donors it is thus more likely that the enhanced wound healing potential that we observed in young BM-MSCs is due to the collective effects of a pro-regenerative phenotype in combination with a larger pool of quiescent MSCs with possibly enhanced stemness. Further studies, e.g. depleting the here identified quiescent MSC subpopulation and variations of knocking down the trophic factors we detected, are suggested to dissect in more detail the functional relevance of these mechanisms in vivo.
CONCLUSION
Our informatics analysis suggests that altered subpopulation dynamics within aged BM-MSCs lead to significant functional deficits. These deficits result in impaired tissue regeneration that can likely be attributed to the loss in trophic potential of these cells. Our work reveals several translational implications: Considering the high incidence of chronic wounds within an increasingly aging population, the application of autologous MSCs to chronic wounds in geriatric patients might be ineffective. Consequentially, young MSC donors are preferable over their aged counterparts in the application of allogeneic regenerative MSC therapies. The observed shift in BM-MSC subpopulation dynamics with a reduction of a highly functional cluster in aged BM-MSCs ultimately suggests a novel pathological mechanism potentially responsible for the numerous deficiencies observed within aged stem and progenitor cells or whole organisms.
Supplementary Material
ACKNOWLEDGMENTS
Flow cytometric analyses for this manuscript were completed at the Stanford Shared Flow Cytometry Facility as well as at the Institute of Clinical and Experimental Transfusion Medicine in Tübingen, Germany with the help of Ursula Hermanutz-Klein. We would also like to thank Yujin Park for expert histology staining, and Gabriele Spohn and Anja Vogel for cell culture. Funding for the stem cell research conducted in our laboratory has been provided by the Hagey Family Endowed Fund in Stem Cell Research and Regenerative Medicine, the Armed Forces Institute of Regenerative Medicine (United States Department of Defense), the National Institutes of Health (R01-DK074095, R01-EB005718, R01-AG025016 to GCG), and the Oak Foundation.
Footnotes
CONFLICT OF INTEREST
The authors do not report any relevant conflicts of interest to disclose.
REFERENCES
- 1.Behrens A, van Deursen JM, Rudolph KL et al. Impact of genomic damage and ageing on stem cell function. Nat Cell Biol. 2014;16:201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Senitz D, Reichenbach A, Smith TG Jr., Surface complexity of human neocortical astrocytic cells: changes with development, aging, and dementia. J Hirnforsch. 1995;36:531–537. [PubMed] [Google Scholar]
- 3.Cieslik KA, Trial J, Crawford JR et al. Adverse fibrosis in the aging heart depends on signaling between myeloid and mesenchymal cells; role of inflammatory fibroblasts. J Mol Cell Cardiol. 2014;70C:56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goichberg P, Kannappan R, Cimini M et al. Age-associated defects in EphA2 signaling impair the migration of human cardiac progenitor cells. Circulation. 2013;128:2211–2223. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Siddiqi S, Sussman MA. Cardiac Hegemony of Senescence. Curr Transl Geriatr Exp Gerontol Rep. 2013;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Despars G, Carbonneau CL, Bardeau P et al. Loss of the osteogenic differentiation potential during senescence is limited to bone progenitor cells and is dependent on p53. PLoS One. 2013;8:e73206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossi DJ, Bryder D, Zahn JM et al. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:9194–9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ting CH, Ho PJ, Yen BL. Age-related decreases of serum-response factor levels in human mesenchymal stem cells are involved in skeletal muscle differentiation and engraftment capacity. Stem Cells Dev. 2014;23:1206–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dudakov JA, Khong DMP, Boyd RL et al. Feeding the fire: the role of defective bone marrow function in exacerbating thymic involution. Trends in Immunology. 2010;31:191–198. [DOI] [PubMed] [Google Scholar]
- 10.Mackall CL, Gress RE. Thymic aging and T-cell regeneration. Immunological Reviews. 1997;160:91–102. [DOI] [PubMed] [Google Scholar]
- 11.Katsimpardi L, Litterman NK, Schein PA et al. Vascular and Neurogenic Rejuvenation of the Aging Mouse Brain by Young Systemic Factors. Science. 2014;344:630–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khong Danika M, Dudakov Jarrod A, Hammett Maree V et al. Enhanced Hematopoietic Stem Cell Function Mediates Immune Regeneration following Sex Steroid Blockade. Stem Cell Reports. 2015;4:445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samse K, Hariharan N, Sussman MA. Personalizing cardiac regenerative therapy: At the heart of Pim1 kinase. Pharmacological Research. 2016;103:13–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai JJ, Dudakov JA, Takahashi K et al. Nrf2 regulates haematopoietic stem cell function. Nat Cell Biol. 2013;15:309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slack JMW. Origin of Stem Cells in Organogenesis. Science. 2008;322:1498–1501. [DOI] [PubMed] [Google Scholar]
- 16.Karp JM, Leng Teo GS. Mesenchymal stem cell homing: the devil is in the details. Cell Stem Cell. 2009;4:206–216. [DOI] [PubMed] [Google Scholar]
- 17.Castillo-Cardiel G, Lopez-Echaury AC, Saucedo-Ortiz JA et al. Bone regeneration in mandibular fractures after the application of autologous mesenchymal stem cells, a randomized clinical trial. Dent Traumatol. 2017;33:38–44. [DOI] [PubMed] [Google Scholar]
- 18.Alvaro-Gracia JM, Jover JA, Garcia-Vicuna R et al. Intravenous administration of expanded allogeneic adipose-derived mesenchymal stem cells in refractory rheumatoid arthritis (Cx611): results of a multicentre, dose escalation, randomised, single-blind, placebo-controlled phase Ib/IIa clinical trial. Ann Rheum Dis. 2017;76:196–202. [DOI] [PubMed] [Google Scholar]
- 19.Aoyama T, Goto K, Kakinoki R et al. An exploratory clinical trial for idiopathic osteonecrosis of femoral head by cultured autologous multipotent mesenchymal stromal cells augmented with vascularized bone grafts. Tissue Eng Part B Rev. 2014;20:233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang YS, Ahn SY, Yoo HS et al. Mesenchymal stem cells for bronchopulmonary dysplasia: phase 1 dose-escalation clinical trial. J Pediatr. 2014;164:966–972 e966. [DOI] [PubMed] [Google Scholar]
- 21.de Oliveira LF, de Oliveira DM, da Silva de Paula LI et al. Transcutaneous parasacral electrical neural stimulation in children with primary monosymptomatic enuresis: a prospective randomized clinical trial. J Urol. 2013;190:1359–1363. [DOI] [PubMed] [Google Scholar]
- 22.Caplan AI, Dennis JE. Mesenchymal stem cells as trophic mediators. Journal of cellular biochemistry. 2006;98:1076–1084. [DOI] [PubMed] [Google Scholar]
- 23.Garg RK, Rennert RC, Duscher D et al. Capillary force seeding of hydrogels for adipose-derived stem cell delivery in wounds. Stem cells translational medicine. 2014;3:1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shabbir A, Cox A, Rodriguez-Menocal L et al. Mesenchymal Stem Cell Exosomes Induce Proliferation and Migration of Normal and Chronic Wound Fibroblasts, and Enhance Angiogenesis In Vitro. Stem Cells Dev. 2015;24:1635–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh L, Brennan TA, Russell E et al. Aging alters bone-fat reciprocity by shifting in vivo mesenchymal precursor cell fate towards an adipogenic lineage. Bone. 2016;85:29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stolzing A, Jones E, McGonagle D et al. Age-related changes in human bone marrow-derived mesenchymal stem cells: Consequences for cell therapies. Mechanisms of Ageing and Development. 2008;129:163–173. [DOI] [PubMed] [Google Scholar]
- 27.Zaim M, Karaman S, Cetin G et al. Donor age and long-term culture affect differentiation and proliferation of human bone marrow mesenchymal stem cells; 2012:1175–1186. [DOI] [PubMed]
- 28.Maredziak M, Marycz K, Tomaszewski KA et al. The Influence of Aging on the Regenerative Potential of Human Adipose Derived Mesenchymal Stem Cells; 2016:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Januszyk M, Gurtner GC. High-Throughput Single-Cell Analysis for Wound Healing Applications. Adv Wound Care (New Rochelle). 2013;2:457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duscher D, Rennert RC, Januszyk M et al. Aging disrupts cell subpopulation dynamics and diminishes the function of mesenchymal stem cells. Scientific Reports. 2014;4:7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kowalczyk MS, Tirosh I, Heckl D et al. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Research. 2015;25:1860–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson Nicola K, Kent David G, Buettner F et al. Combined Single-Cell Functional and Gene Expression Analysis Resolves Heterogeneity within Stem Cell Populations. Cell Stem Cell.16:712–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glotzbach JP, Januszyk M, Vial IN et al. An information theoretic, microfluidic-based single cell analysis permits identification of subpopulations among putatively homogeneous stem cells. PLoS One. 2011;6:e21211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rennert RC, Januszyk M, Sorkin M et al. Microfluidic single-cell transcriptional analysis rationally identifies novel surface marker profiles to enhance cell-based therapies. Nature communications. 2016;7:11945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.