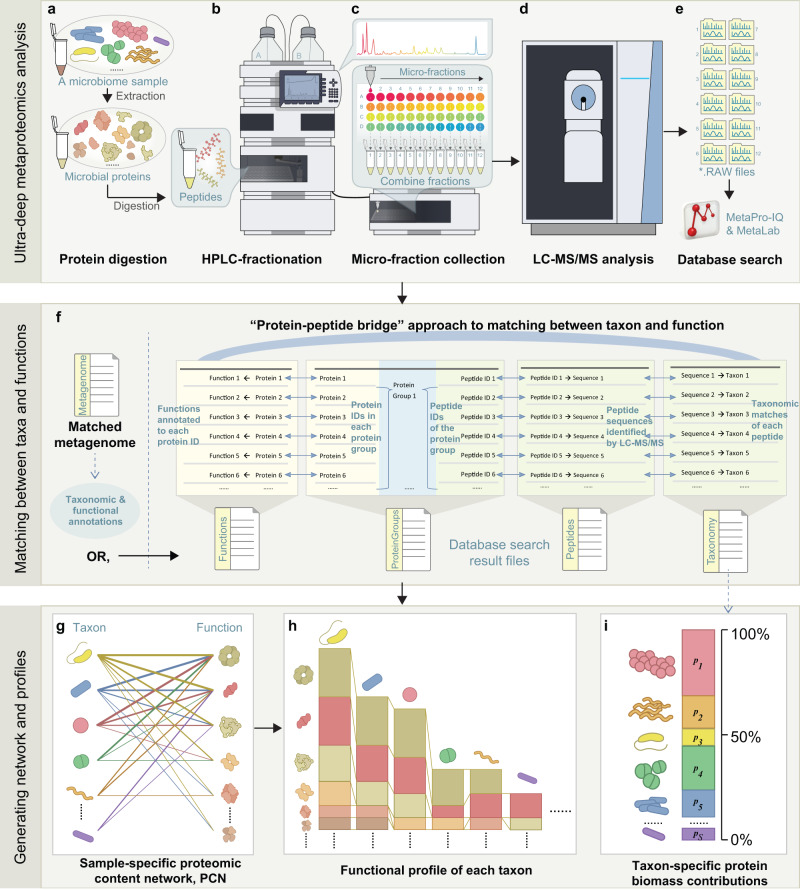
Fig. 1. Computation of within-sample proteomic-content network and functional, taxonomic profiles using an ultra-deep metaproteomics approach.
a Each individual’s gut microbiome sample was subjected to protein extraction. Then, purified proteins were digested by trypsin. b The resulting peptides were fractionated using a high-pH reversed-phase approach. c 48 micro-fractions were combined into 12 samples prior to LC-MS/MS analysis. d In this study, LC-MS/MS analysis was performed using Exploris 480 with a 1 h gradient for each fraction. e The LC-MS/MS *.RAW files were searched against a protein database using MetaPro-IQ workflow and MetaLab. f When matched metagenomes are available, taxonomic and functional annotations of the metagenomes can be used for PCN generation. Alternatively, a protein-peptide bridge approach can be used for generating the PCNs from MetaLab result tables (see details in Methods and Supplementary Notes 1) without the need of matched metagenomes. g The sample-specific PCN is a bipartite plot linking taxon to their expressed functions, with the links weighed by protein abundances of each taxon. h The PCN can also be converted as functional profiles of each taxon. i The taxon-specific protein biomass contributions (percentage) were calculated from the taxonomy table based on summing up unique peptides of each taxon.