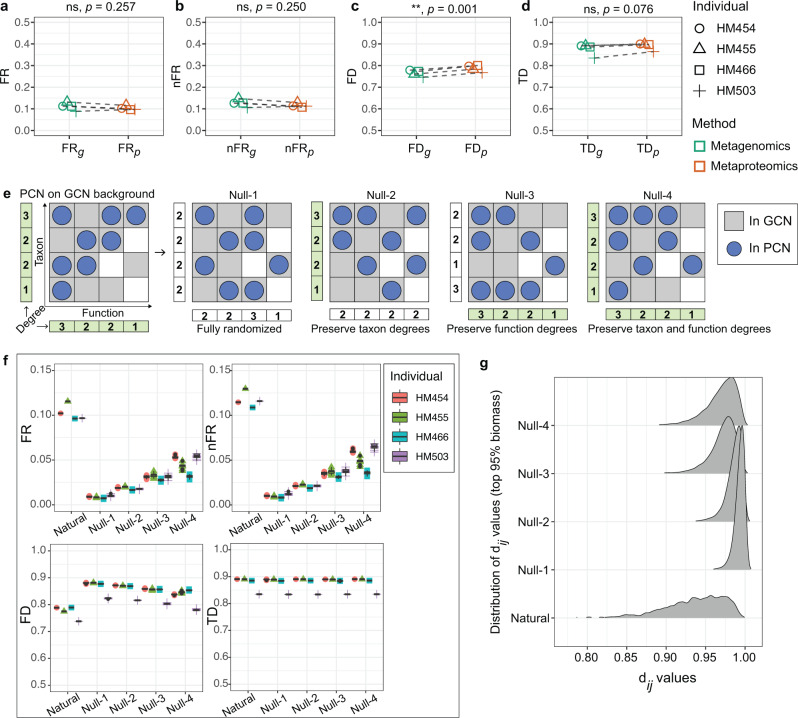
Fig. 4. Naturally occurring proteomic-content networks contribute to high functional redundancy in human gut microbiomes.
a Within-sample functional redundancy (FR) in the metagenomes versus in the metaproteomes of the individual microbiomes. b Within-sample FR normalized by taxonomic diversity (nFR) in the metagenomes versus in the metaproteomes of the individual microbiomes. c Functional diversity (FD) in the metagenomes versus in the metaproteomes. d Taxonomic diversity (TD) in the metagenomes versus in the metaproteomes. For (a–d), paired t-tests (two-sided) were performed, ** indicates statistical significance at the p < 0.01 level. e Scheme diagram showing the creation of four null networks by randomizing a naturally occurred PCN under its corresponding GCN background. Null-1 PCN is a completely randomized network under the GCN background, Null-2 PCN preserves the taxon degrees, Null-3 PCN preserves the function degrees, and Null-4 PCN preserves both taxon and function degrees. f Comparison of FRp, nFRp, FDp and TDp among naturally occurring PCN and four types of null PCNs in the four individuals. Each of the four null networks was randomized 10 times to demonstrate the reproducibility of the results. Lower and upper hinges correspond to the first and third quartiles, thick line in the box corresponds to the median, and whiskers represent the maximum and minimum, excluding outliers. g Distribution of dij values in natural PCNs and Null-1 ~ 4 PCNs. dij distributions of genera making up the top 95% protein biomass of the dataset are shown. Source data are provided as a Source Data file.